Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 13
Bioinformatics-based identification of potential microRNA biomarkers in frequent and non-frequent exacerbators of COPD
Authors Liu X , Qu J, Xue W, He L, Wang J, Xi X, Liu X, Yin Y , Qu Y
Received 23 January 2018
Accepted for publication 2 March 2018
Published 16 April 2018 Volume 2018:13 Pages 1217—1228
DOI https://doi.org/10.2147/COPD.S163459
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chunxue Bai
Xiao Liu,1,* Jingge Qu,2,* Weixiao Xue,1 Liangai He,1 Jun Wang,3 Xuejiao Xi,1 Xiaoxia Liu,1 Yunhong Yin,1 Yiqing Qu1
1Department of Respiratory Medicine, Qilu Hospital of Shandong University, Jinan, People’s Republic of China; 2Department of Rheumatology, Second Hospital of Harbin Medical University, Harbin, People’s Republic of China; 3Department of Respiratory Medicine, Second Hospital of Shandong Traditional Chinese Medicine University, Jinan, People’s Republic of China
*These authors contributed equally to this work
Objectives: MicroRNAs (miRNAs) play essential roles in the development of COPD. In this study, we aimed to identify and validate potential miRNA biomarkers in frequent and non-frequent exacerbators of COPD patients using bioinformatic analysis.
Materials and methods: The candidate miRNA biomarkers in COPD were screened from Gene Expression Omnibus (GEO) dataset and identified using GEO2R online tool. Then, we performed bioinformatic analyses including target prediction, gene ontology (GO), pathway enrichment analysis and construction of protein–protein interaction (PPI) network. Furthermore, the expression of the identified miRNAs in peripheral blood monocular cells (PBMCs) of COPD patients was validated using quantitative real-time polymerase chain reaction (qRT-PCR).
Results: MiR-23a, miR-25, miR-145 and miR-224 were identified to be significantly downregulated in COPD patients compared with healthy controls. GO analysis showed the four miRNAs involved in apoptotic, cell differentiation, cell proliferation and innate immune response. Pathway analysis showed that the targets of these miRNAs were associated with p53, TGF-β, Wnt, VEGF and MAPK signal pathway. In healthy controls, the miR-25 and miR-224 levels were significantly decreased in smokers compared with nonsmokers (P<0.001 and P<0.05, respectively). In COPD patients, the levels of miR-23a, miR-25, miR-145 and miR-224 were associated with Global Initiative for Chronic Obstructive Lung Disease (GOLD) stages. Notably, miR-23a and miR-145 were significantly elevated in non-frequent exacerbators compared with frequent exacerbators (P<0.05), and miR-23a showed higher area under the receiver–operator characteristic curve (AUROC) than miR-145 (0.707 vs 0.665, P<0.05).
Conclusion: MiR-23a, miR-25, miR-145 and miR-224 were associated with the development of COPD, and miR-23a might be a potential biomarker for discriminating the frequent exacerbators from non-frequent exacerbators.
Keywords: COPD, microRNAs, bioinformatic analysis, Gene Expression Omnibus dataset, biomarkers, Global Initiative for Chronic Obstructive Lung Disease stages
Introduction
COPD is characterized by persistent airflow limitation due to airway and/or alveolar abnormalities that are usually caused by exogenous exposure to noxious particles or gases.1 Cigarette smoking is the major risk factor for the development of COPD. In 2010, COPD is the fourth leading cause of death worldwide, but it is predicted to be the third by 2020.2 Acute exacerbation of COPD (AECOPD), which is generally characterized by the aggravated dyspnea and the increased volumes of phlegm and phlegm purulence, had an in-hospital mortality of 4.3%–7.7%.3–5 However, there are still no strong and effective strategies to predict the occurrence of AECOPD. Therefore, biomarkers for predicting the development and prognosis of COPD are urgently required.
MicroRNAs (miRNAs) are endogenous, approximately 20–25 nt, noncoding RNAs that can target mRNAs for the cleavage of translational repression.6 miRNAs have been demonstrated to participate in the pathogenesis and development of COPD. Fujita et al7 reported that miR-210 contributed to abnormal airway remodeling by suppressing autophagy and promoting myofibroblast differentiation. Osei et al8 demonstrated that miR-146a-5p involved in the epithelial–fibroblast cross talk and the blockage of miR-146a-5p might induce the chronic inflammation in COPD-derived primary human lung fibroblasts. Furthermore, miRNAs have also been demonstrated to be promising and effective biomarkers for the diagnosis of COPD. In 2012, Akbas et al9 first reported the downregulation of serum miR-20, miR-28-3p, miR-34c-5p and miR-100 and the upregulation of miR-7 in COPD patients compared with healthy controls, and they suggested that these miRNAs might provide potential biomarkers for therapeutic strategy. Another group reported that the ratio of serum miR-20a to miR-181a was associated with the early stage of COPD in asymptomatic heavy smokers.10 In addition, Wang et al11 also demonstrated that the levels of miR-145-5p, miR-338-3p and miR-3620-3p were related to the severity of COPD. However, these preliminary results were usually limited by small sample size and have not been confirmed using large-scale dataset.
With the development of high-throughput microarray and sequencing technology, several public resources have been established, and the Gene Expression Omnibus (GEO) of National Center for Biotechnology Information (NCBI) is the largest one. Bioinformatic analyses based on the GEO database provide valuable information for searching biomarkers in various diseases.12–16 However, little data have been reported on the bioinformatics-based identification of potential miRNA biomarkers for COPD patients. Therefore, we aimed to identify and validate potential miRNA biomarkers in frequent and non-frequent exacerbators of COPD patients using bioinformatic analysis.
Materials and methods
Selection of candidate miRNAs and target prediction
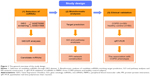
The general overview of the study design is shown in Figure 1. Shared blood miRNA profiles of COPD patients were searched in GEO datasets.17 GSE70080 and GSE31568 were selected, and the dysregulated miRNAs of COPD patients and healthy controls were intersected. GEO2R18 was performed to compare the original submitter-supplied processed data tables using the GEO query and limma R packages from the bioconductor project. Generally, GEO2R is a well-designed tool for avoiding the false positivity by handling the various types of data and performing multiple testing corrections on P-values.
The accuracy for the predicted targets of candidate miRNAs was improved by the following software: PicTar19, miRDB20 and TargetScan.21 Validated targets were acquired from TarBase.22 Briefly, we took the intersection of the three series of predicted targets and validated the targets using TarBase.
Gene ontology (GO) and pathway enrichment analysis of target genes
GeneCodis23 is a web-based tool for comprehensive analysis of gene annotations, encompassing GO, Panther and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, InterPro Motifs and Transcription Factors, etc. GO annotates genes and gene products including molecular function (MF), cellular component (CC) and biological process (BP).24 Panther and KEGG are bioinformatic resources for genomic and gene functional information.25,26 The significant enrichment results were accepted at a threshold of ≥2 gene counts with a P-value <0.05.
Construction of protein–protein interaction (PPI) network
Venn diagram was carried out for finding potential targets of the candidate miRNAs (Venny 2.1).27 The interactions of the potential target genes were predicted by the Search Tool for the Retrieval of Interacting Genes (STRING) database.28 Cytoscape (version 3.5.1; http://cytoscape.org/) is an open source software which is capable of integrating the high-throughput data and other types of molecular data into a unitized conceptual framework.29 The values of gene interactions predicted by STRING were imported into Cytoscape to visualize the resulting PPIs and to identify hub genes among potential targets.
Patients and healthy controls
A total of consecutive 50 COPD patients were included during October 2016 to February 2017 in Qilu Hospital of Shandong University, as well as 50 well age- and gender-matched healthy subjects were recognized as controls. The inclusive criteria for COPD patients were set as forced expiratory volume in 1 s/forced vital capacity (FEV1/FVC)<0.7 according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) guideline. The classification of the severity stage for COPD was the following: GOLD I, forced expiratory volume in the first second of expiration for predicted values (FEV1%pre)≥80%; GOLD II, 50%≤FEV1%pre<80%; GOLD III, 30%≤FEV1%pre<50%; GOLD IV, FEV1%pre<30%.1 Frequent exacerbator was identified as COPD patient with two or more episodes of acute exacerbations during preceding 1 year, while non-frequent exacerbator as COPD patient with <2 episodes of acute exacerbations during preceding 1 year. Meanwhile, all patients did not undergo acute exacerbation during the nearly past 3 months. Exclusion criteria included the existences of other chronic lung diseases, tumors, unstable cardiovascular diseases, diabetes, nervous system diseases and liver and kidney diseases. The study was approved by the medical ethics committee of Qilu Hospital of Shandong University, and the written informed consent form was obtained from each patient.
Peripheral blood mononuclear cell (PBMC) separation and RNA extraction
The fasting peripheral vein blood (4 mL) was collected in EDTA tubes, and PBMCs were obtained through the Ficoll (Solarbio Life Sciences, Beijing, People’s Republic of China) isolation method. Total RNA containing small RNA was extracted from PBMCs by using the Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. RNA purity and concentration were evaluated by the ratio of absorbance at OD260/OD280 through ultraviolet spectrophotometer.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with Bulge-loop™ miRNA qRT-PCR Starter kit on RT-PCR system (Thermo Fisher Scientific) according to the following protocol: reverse transcription PCR: 42°C 60 min, 70°C 10 min, 4°C ∞; qPCR: 95°C for 10 min, followed by 40 cycles of 95°C for 2 s, 60°C for 20 s and then 70°C for 10 s. Melt curve analysis was performed; the nuclear acid melting temperatures were 70°C–95°C; the heating rate was 0.5°C/time; holding time was 5 s/time; then, the Ct was recorded. The primers of these miRNAs and U6 were obtained from RiboBio Corporation (Guangzhou, People’s Republic of China). The fold change of each miRNA expression was calculated using the ΔΔCT method.
Statistical analyses
The demography and clinical data are presented as mean ± SD. The difference of gender composition between healthy controls and COPD patients was analyzed by Pearson’s chi-square test, and the differences for continuous variables were analyzed by Mann–Whitney U-test. The relationship between FEV1%pre and pack-years of COPD patients was analyzed by Pearson’s correlation analysis. The Mann–Whitney U-test was employed to compare the expression levels of miRNAs between the two groups. Receiver–operator characteristic (ROC) curve analysis and the area under the ROC curve (AUROC) were used to evaluate the specificity and sensitivity of miRNAs, with the larger AUROC value showing the higher diagnostic values. All statistical analyses were performed using the SPSS 20.0 software (IBM Corporation, Armonk, NY, USA) and the GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Differences with P-value of <0.05 were considered statistically significant.
Results
Identification of candidate miRNAs and prediction of targets
In GSE31568 and GSE70080, COPD samples and healthy controls were entered into GEO2R analysis to identify the candidate miRNAs. MiR-23a, miR-25, miR-145 and miR-224 have been selected as candidate miRNAs (P<0.05). The target genes were predicted from both prediction algorithms (TargetScan, miRDB and PicTar) and experimentally supported targets from TarBase. Eventually, we gained a total of 3114 target genes of the candidate miRNAs.
GO, KEGG and Panther pathway functional analyses
GO enrichment analysis demonstrated that the main annotations for the MF of the 3114 target genes included DNA-dependent regulation of transcription, signal transduction, multicellular organismal development, apoptotic process, transmembrane transport, cell differentiation, cell proliferation, innate immune response, platelet activation, response to stress, response to hypoxia, cell adhesion, metabolic process, cell migration, lipid metabolic process, oxidation–reduction process, inflammatory response, cytokine-mediated signaling pathway, etc (Figure 2A).
KEGG and Panther pathway analysis of these target genes presented p53 signal pathway, TGF-β signal pathway, Wnt signal pathway, VEGF signal pathway, MAPK signal pathway, oxidative stress response, Notch signal pathway, etc, indicating their potential roles for the pathogenesis of COPD (Figure 2B and C). The most enriched items of KGEE and Panther pathways according to P-values are presented in Table 1.
PPI network construction of potential targets
The Venn diagram of 3114 predicted genes is shown in Figure 2D. There are 32 common target genes (FBN1, ADD3, AHNAK, AP1G1, BAZ2A, etc) for miR-23a, miR-25 and miR-145, four genes (BTG2, TMEM9B, ELK4, ACTB) for miR-25, miR-145 and miR-224, 27 genes (MARCKS, YWHAG, SLC4A4, ARFGEF2, BIR3BP, etc) for miR-23, miR-25, miR-224, three genes (ZNF423, KDM5A, USP37) for miR-23, miR-145 and miR-224, as well as six genes (CCNG1, CCND1, CELF1, DDX3X, MAP1B and NUFIP1) for all the four miRNAs. Furthermore, the PPI network of the 72 identified target genes is shown in Figure 3. In Figure 3, ACTB, MYC, CTNNB1, CDH1 and CCND1 are shown as hub target genes according to the counts of interacting protein.
Basic demographic characteristics of the COPD patients and healthy controls
In this current study, the validation set included a total of 50 COPD patients (four for GOLD I, 14 for GOLD II, 15 for GOLD III and 17 for GOLD IV) and 50 healthy controls (32 nonsmokers and 18 smokers). The demographic and clinical characteristics of COPD patients and healthy controls are presented in Tables 2 and 3. The post-bronchodilator (BD) FEV1%pre of each GOLD grade (mean ± SD) is shown as follows: GOLD I, 86.29%±4.63%; GOLD II, 56.47%±6.51%; GOLD III, 38.74%±5.44%; GOLD IV, 19.83%±7.03%. In COPD patients, there were 18 non-frequent exacerbators and 32 frequent exacerbators. FEV1%pre was demonstrated to be negatively correlated with pack-years in COPD patients (P<0.05, r=−0.298; Figure 4). Between healthy controls and COPD patients, there were no significant differences in age and body mass index (BMI; all P>0.05) as well as significant difference in smoking status (P<0.001).
Validation of the identified miRNAs in PBMCs of healthy and COPD subjects
The levels of miR-23a, miR-25, miR-145 and miR-224 in PBMCs were found to be significantly lower in COPD patients than those in healthy controls (Figure 5A). Interestingly, miR-23a and miR-145 were downregulated in frequent exacerbators compared with non-frequent exacerbators (P<0.05). In healthy controls, the levels of miR-25 and miR-224 were upregulated in nonsmokers compared with those in smokers (P<0.001, P<0.05, respectively).
As shown in Figure 5B, the levels of miR-23a and miR-145 in GOLD I and GOLD II were significantly higher than GOLD III (P<0.05) and GOLD IV (P<0.001), as well as the same trend of miR-23a and miR-145 in GOLD III compared with GOLD IV (P<0.05). There was no significant difference in miR-25 between GOLD I and GOLD II compared with GOLD III (P>0.05), while there was a significantly higher expression of miR-25 in GOLD I and GOLD II and GOLD III compared with GOLD IV (P<0.01 and P<0.05, respectively). For the level of miR-224, there was no significant difference in GOLD I and GOLD II compared with GOLD III (P>0.05), as well as the same trend between GOLD III and GOLD IV (P>0.05). Of note, we demonstrated the significant higher expression of miR-224 in GOLD I than GOLD IV (P<0.01).
ROC curve analyses of miR-23a and miR-145 as biomarkers in non-frequent and frequent COPD exacerbators
ROC analysis was performed to estimate the diagnostic accuracy of miR-23a and miR-145 as biomarkers for identifying frequent and non-frequent exacerbators (Figure 6). The AUROC was 0.707 (95% CI: 0.543–0.880) with a sensitivity of 71.88% and specificity of 66.67% for miR-23a (P=0.016), as well as 0.665 (95% CI: 0.5123–0.8175) with a sensitivity of 65.63% and specificity of 66.67% for miR-145 (P=0.066).
Discussion
AECOPD is an acute event of respiratory symptoms with serious impact on quality of life of COPD patients.30 However, little data have been reported on exploring the miRNA biomarkers for identifying frequent and non-frequent exacerbators of COPD patients. In the current study, we first identified the profile of miR-23a, miR-25, miR-145 and miR-224 and provided the functional pathways for those miRNAs using bioinformatic methods. Furthermore, we demonstrated that miR-23a and miR-145 were significantly different in non-frequent and frequent exacerbators using validated cohort. Finally, we suggested that miR-23a might be the potential and promising biomarker for discriminating frequent exacerbators from non-frequent exacerbators of COPD patients.
In recent years, increasing evidences showed that miRNAs contributed to the development and progression of COPD patients. MiR-199a-5p was demonstrated to be a critical regulator for unfolded protein response in alpha-1 antitrypsin-deficient monocytes.31 Blockage of miR-503 has been reported to augment the release of vascular endothelial growth factor from lung fibroblasts of COPD patients.32 MiR-218-5p, which was significantly downregulated in COPD patients compared with nonsmokers, was recognized as a protective factor for COPD.33 Furthermore, Shen et al34 reported that miR-483-5p was capable of abrogating the TGF-β-mediated cell proliferation in pulmonary epithelial and fibroblast cell lines. In addition, Shen et al35 demonstrated that miR-149-3p might attenuate the inflammatory response via TLR-4/NF-κB signaling pathway in COPD patients.
In the current study, we first identified that the profiles of miR-23a, miR-25, miR-145 and miR-224 were downregulated in COPD patients than healthy controls, indicating their potential roles in the development of COPD. Previously, miR-23a, miR-25, miR-145 and miR-224 have been found to be abnormally expressed in idiopathic pulmonary hypertension,36 asthma37,38 and lung cancer.39–41 MiR-23a was reported to be associated with the loss of muscle force during the AECOPD,42 and the mechanism might be that miR-23a could regulate TLR2/MyD88/NF-κB pathway by targeting TLRs.43 MiR-25 was demonstrated to mediate the phenotype of human airway smooth muscle cells (ASMCs) when exposing to IL-1β, TNF-α and IFN-γ.44 MiR-145 can modulate the Th1/Th2 balance in asthma by regulating Runx3.37 In the study by Hu et al,40 miR-145 inhibited the TGF-β-induced epithelial–mesenchymal transition (EMT) by negatively regulating SMAD3 expression in human non-small cell lung cancer (NSCLC) cells. MiR-224 was reported to inhibit cell proliferation and metastasis in NSCLC A549 cell lines.41 Shen et al45 reported that miR-224 could downregulate gene expression of ACADM and ALDH2, inhibit the formation of triglyceride and influence lipid metabolism and apoptosis. Therefore, it is hypothesized that the profiles of miR-23a, miR-25, miR-145 and miR-224 might be associated with the inflammation, oxidative stress, immune imbalance, EMT, cell proliferation, apoptosis and lipid metabolism of COPD.
We have performed the GO and pathway analysis to predict the possible biological function and signal pathways of the target genes in the pathogenesis of COPD. Our results demonstrated that miR-23a, miR-25, miR-145 and miR-224 may play important roles in inflammation, oxidative stress, lipid metabolic disorders, cell proliferation and apoptosis. Enriched pathways included p53 signal pathway, TGF-β signal pathway, Wnt signal pathway, VEGF signal pathway, MAPK signal pathway, oxidative stress and Notch signal pathway. Tilley et al46 reported the lower expression of NOTCH3 gene in smokers than nonsmokers, suggesting its potential interaction with smoke. Besides, TGF-β signal pathway involved in the development of lung, inflammation and airway remodeling of COPD. Another study showed that smoking-mediated airway inflammation could lead to oxidative stress and airway remodeling by activating MMP-9 and TGF-β/Smads pathway.47 Therefore, we believe that our identified candidate miRNAs might contribute to the inflammation, airway mucus hypersecretion, oxidative stress and airway remodeling via those enriched pathways. However, the hypothesis based on the bioinformatic analyses is urgently needed to be confirmed in the future study.
In the current study, GSE70080 and GSE31568 were selected to perform the differential miRNA expression profile of COPD patients and healthy controls. It is worth noting that the two GEO Series also contain the gene information of lung cancer.48,49 Using the same GSE datasets, Halvorsen et al48 revealed six circulating miRNAs (miR-429, miR-205, miR-200b, miR-203, miR-125b and miR-34b) for the early diagnosis of NSCLC patients, and Keller et al49 reported some potential miRNA biomarkers for distinguishing lung cancer from COPD. Our current study mainly compared the data from COPD patients and healthy controls and demonstrated that miR-23a, miR-25, miR-145 and miR-224 were associated with the development of COPD.
Frequent exacerbation phenotype is one of the main phenotypes in COPD patients,50,51 and biomarkers for identifying the frequent and non-frequent exacerbators are urgently needed. In the previous study, urinary desmosine was reported to be a useful biomarker for identifying frequent exacerbators of COPD patients.52 Besides, the upregulation of IL-1 pathway mediators (IRAK2, IRAK3, PELI1 and IL1R1)53 was reported to be relevant to frequent exacerbations of COPD. Fu et al54 demonstrated that serum IL-6 and sputum IL-1β were associated with the frequent exacerbations. However, miRNA biomarkers for identifying the frequent exacerbators of COPD have not been reported to date. Using the clinical samples, we demonstrated that miR-23a and miR-145 were significantly expressed between non-frequent and frequent exacerbators of COPD. Furthermore, ROC curve showed that miR-23a might be the potential and promising biomarker for discriminating frequent exacerbators from non-frequent exacerbators.
However, there are some limitations in our study. First, we performed bioinformatic analysis using two different groups and platforms, which might generate variances. Usually, there are two commonsense methods to avoid the variances: the first method is for performing quality control within each group using the techniques of normalization, principal component analysis (PCA) and cluster, then merging all the groups with raw data after quality control and eventually performing differential expression profiling in the merged group.55–58 However, the quality control might be quite disordered especially for the dataset with relatively small samples and inevitably generate poor repeatability. The second method is for first performing differential expression profiling in each group, respectively, and then choosing the common intersection. The P-value would be assigned for all miRNAs in the ranked lists to re-rank these miRNAs and decide their significance.59–64 This method might be more feasible for the dataset with relatively small samples or platform heterogeneity. Therefore, considering the real spirit of our current study, we have selected the second method to avoid the variances. Second, we still need to investigate the functional mechanism of the target genes and pathways in the future study, and the prognostic value of miR-23a should be confirmed in the prospective, multicenter, large-scale cohort.
Conclusion
We first identified the profile of miR-23a, miR-25, miR-145 and miR-224 and provided the functional pathways for those miRNAs using bioinformatic methods. Furthermore, we demonstrated that miR-23a and miR-145 were significantly different in non-frequent and frequent exacerbators using validated cohort. Finally, we suggested that miR-23a might be the potential and promising biomarker for discriminating frequent exacerbators from non-frequent exacerbators of COPD patients. However, the functional mechanism of the target genes and the prognostic value of miR-23a should be confirmed in the future study.
Acknowledgments
This study was supported by the grants from the Science and Technology Foundation of Shandong Province (grant number: 2007GG3002008, 2015GGH318002 and 2015GSF118064). Xiao Liu and Jingge Qu are the co-first authors.
Disclosure
The authors report no conflicts of interest in this work.
References
Global Strategy for the Diagnosis, Management and Prevention of COPD, Global Initiative for Chronic Obstructive Lung Diseases (GOLD) 2016. Available from: http://goldcopd.org/. Accessed Octobor 20, 2017. | ||
Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. | ||
Perera PN, Armstrong EP, Sherrill DL, Skrepnek GH. Acute exacerbations of COPD in the United States: inpatient burden and predictors of costs and mortality. COPD. 2012;9(2):131–141. | ||
Johannesdottir SA, Christiansen CF, Johansen MB, et al. Hospitalization with acute exacerbation of chronic obstructive pulmonary disease and associated health resource utilization: a population-based Danish cohort study. J Med Econ. 2013;16(7):897–906. | ||
de Miguel-Diez J, Jimenez-Garcia R, Hernandez-Barrera V, et al. Trends in hospital admissions for acute exacerbation of COPD in Spain from 2006 to 2010. Respir Med. 2013;107(5):717–723. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Fujita Y, Araya J, Ito S, et al. Suppression of autophagy by extracellular vesicles promotes myofibroblast differentiation in COPD pathogenesis. J Extracell Vesicles. 2015;4:28388. | ||
Osei ET, Florez-Sampedro L, Tasena H, et al. miR-146a-5p plays an essential role in the aberrant epithelial-fibroblast cross-talk in COPD. Eur Respir J. 2017;49(5):1602538. | ||
Akbas F, Coskunpinar E, Aynaci E, Oltulu YM, Yildiz P. Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp Lung Res. 2012;38(6):286–294. | ||
Xie L, Wu M, Lin H, et al. An increased ratio of serum miR-21 to miR-181a levels is associated with the early pathogenic process of chronic obstructive pulmonary disease in asymptomatic heavy smokers. Mol Biosyst. 2014;10(5):1072–1081. | ||
Wang M, Huang Y, Liang Z, et al. Plasma miRNAs might be promising biomarkers of chronic obstructive pulmonary disease. Clin Respir J. 2016;10(1):104–111. | ||
Wang Y, Xue D, Li Y, et al. The long noncoding RNA MALAT-1 is a novel biomarker in various cancers: a meta-analysis based on the GEO database and literature. J Cancer. 2016;7(8):991–1001. | ||
Huang Y, Zhu J, Li W, et al. Serum microRNA panel excavated by machine learning as a potential biomarker for the detection of gastric cancer. Oncol Rep. 2018;39(3):1338–1346. | ||
Hu T, Xi J. Identification of COX5B as a novel biomarker in high-grade glioma patients. Onco Targets Ther. 2017;10:5463–5470. | ||
Tian X, Zhu X, Yan T, et al. Differentially expressed lncRNAs in gastric cancer patients: a potential biomarker for gastric cancer prognosis. J Cancer. 2017;8(13):2575–2586. | ||
Manchia M, Piras IS, Huentelman MJ, et al. Pattern of gene expression in different stages of schizophrenia: down-regulation of NPTX2 gene revealed by a meta-analysis of microarray datasets. Eur Neuropsychopharmacol. 2017;27(10):1054–1063. | ||
Barrett T, Suzek TO, Troup DB, et al. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33 (Database issue):D562–D526. | ||
Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. | ||
Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. | ||
Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32(9):1316–1322. | ||
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. | ||
Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. | ||
Nogales-Cadenas R, Carmona-Saez P, Vazquez M, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37(Web Server issue):W317–W322. | ||
Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(Database issue):D322–D326. | ||
Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38(Database issue):D204–D210. | ||
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. | ||
Oliveros JC. (2007–2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. Available from: http://bioinfogp.cnb.csic.es/tools/venny/index.html | ||
Jensen LJ, Kuhn M, Stark M, et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucl Acids Res. 2009;37(Database issue):D412–D416. | ||
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. | ||
Wedzicha JA, Seemungal TA. COPD exacerbations: defining their cause and prevention. Lancet. 2007;370(9589):786–796. | ||
Hassan T, Carroll TP, Buckley PG, et al. miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and alpha1-antitrypsin deficiency. Am J Respir Crit Care Med. 2014;189(3):263–273. | ||
Ikari J, Nelson AJ, Obaid J, et al. Reduced microRNA-503 expression augments lung fibroblast VEGF production in chronic obstructive pulmonary disease. PLoS One. 2017;12(9):e0184039. | ||
Conickx G, Mestdagh P, Avila Cobos F, et al. MicroRNA profiling reveals a role for microRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195(1):43–56. | ||
Shen Z, Tang W, Guo J, Sun S. miR-483-5p plays a protective role in chronic obstructive pulmonary disease. Int J Mol Med. 2017;40(1):193–200. | ||
Shen W, Liu J, Zhao G, et al. Repression of Toll-like receptor-4 by microRNA-149-3p is associated with smoking-related COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:705–715. | ||
Sarrion I, Milian L, Juan G, et al. Role of circulating miRNAs as biomarkers in idiopathic pulmonary arterial hypertension: possible relevance of miR-23a. Oxid Med Cell Longev. 2015;2015:792846. | ||
Qiu YY, Zhang YW, Qian XF, Bian T. miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am J Trans Res. 2017;9(7):3184–3199. | ||
Suojalehto H, Lindstrom I, Majuri ML, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014;163(3):168–178. | ||
Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–4942. | ||
Hu H, Xu Z, Li C, et al. MiR-145 and miR-203 represses TGF-beta-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer. 2016;97:87–94. | ||
Zhu D, Chen H, Yang X, et al. Decreased microRNA-224 and its clinical significance in non-small cell lung cancer patients. Diagn Pathol. 2014;9:198. | ||
Duan Y, Zhou M, Xiao J, et al. Prediction of key genes and miRNAs responsible for loss of muscle force in patients during an acute exacerbation of chronic obstructive pulmonary disease. Int J Mol Med. 2016;38(5):1450–1462. | ||
Gu X, Gao Y, Mu DG, Fu EQ. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-kappaB pathway by targeting TLR2. Exp Cell Res. 2017;354(2):71–77. | ||
Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42(4):506–513. | ||
Shen B, Pan Q, Yang Y, et al. miR-224 affects mammary epithelial cell apoptosis and triglyceride production by downregulating ACADM and ALDH2 genes. DNA Cell Biol. 2017;36(1):26–33. | ||
Tilley AE, Harvey BG, Heguy A, et al. Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179(6):457–466. | ||
Churg A, Cosio M, Wright JL. Mechanisms of cigarette smoke-induced COPD: insights from animal models. Am J Physiol Lung Cell Mol Physiol. 2008;294(4):L612–L631. | ||
Halvorsen AR, Bjaanaes M, LeBlanc M, et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget. 2016;7(24):37250–37259. | ||
Keller A, Leidinger P, Bauer A, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8(10):841–843. | ||
Hurst JR, Vestbo J, Anzueto A, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363(12):1128–1138. | ||
Tashkin DP. Frequent exacerbations of chronic obstructive pulmonary disease – a distinct phenotype? N Engl J Med. 2010;363(12):1183–1184. | ||
Kim C, Ko Y, Kim SH, et al. Urinary desmosine is associated with emphysema severity and frequent exacerbation in patients with COPD. Respirology. 2018;23(2):176–181. | ||
Baines KJ, Fu JJ, McDonald VM, Gibson PG. Airway gene expression of IL-1 pathway mediators predicts exacerbation risk in obstructive airway disease. Int J Chron Obstruct Pulmon Dis. 2017;12:541–550. | ||
Fu JJ, McDonald VM, Baines KJ, Gibson PG. Airway IL-1beta and Systemic Inflammation as Predictors of Future Exacerbation Risk in Asthma and COPD. Chest. 2015;148(3):618–629. | ||
Shangguan H, Tan SY, Zhang JR. Bioinformatics analysis of gene expression profiles in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2015;19(11):2054–2061. | ||
Kong XD, Liu N, Xu XJ. Bioinformatics analysis of biomarkers and transcriptional factor motifs in Down syndrome. Braz J Med Biol Res. 2014;47(10):834–841. | ||
Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017;18(4):E722. | ||
Park C, Yu N, Choi I, Kim W, Lee S. lncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics. 2014;30(17):2480–2485. | ||
Song E, Song W, Ren M, et al. Identification of potential crucial genes associated with carcinogenesis of clear cell renal cell carcinoma. J Cell Biochem. Epub 2017 Dec 11. | ||
Song YJ, Li G, He JH, Guo Y, Yang L. Bioinformatics-based identification of microRNA-regulated and rheumatoid arthritis-associated genes. PLoS One. 2015;10(9):e0137551. | ||
Zheng LQ, Wang R, Chi SM, Li CX. Matrix metalloproteinase 1: a better biomarker for squamous cell carcinoma by multiple microarray analyses. G Ital Dermatol Venereol. Epub 2017 Dec 15. | ||
Li H, Wang H, Sun Z, Guo Q, Shi H, Jia Y. The clinical and prognostic value of polo-like kinase 1 in lung squamous cell carcinoma patients: immunohistochemical analysis. Biosci Rep. Epub 2017 Jul 19. | ||
Lin ZF, Shen XY, Lu FZ, Ruan Z, Huang HL, Zhen J. Reveals new lung adenocarcinoma cancer genes based on gene expression. Eur Rev Med Pharmacol Sci. 2012;16(9):1249–1256. | ||
Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. Differences in gene expression profiles and carcinogenesis pathways involved in cisplatin resistance of four types of cancer. Oncol Rep. 2013;30(2):596–614. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.