Back to Journals » International Journal of General Medicine » Volume 15
Bioinformatics Analysis of the Key Genes and Pathways in Multiple Myeloma
Authors Sheng X, Wang S, Huang M, Fan K, Wang J, Lu Q
Received 9 June 2022
Accepted for publication 19 August 2022
Published 5 September 2022 Volume 2022:15 Pages 6999—7016
DOI https://doi.org/10.2147/IJGM.S377321
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xinge Sheng,1,2,* Shuo Wang,2,* Meijiao Huang,1 Kaiwen Fan,1,2 Jiaqi Wang,1,2 Quanyi Lu1,2
1Department of Hematology, Zhongshan Hospital Xiamen University, Xiamen, People’s Republic of China; 2Clinical Medicine Department, School of Medicine, Xiamen University, Xiamen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Quanyi Lu, Tel +86 13600959425, Email [email protected]
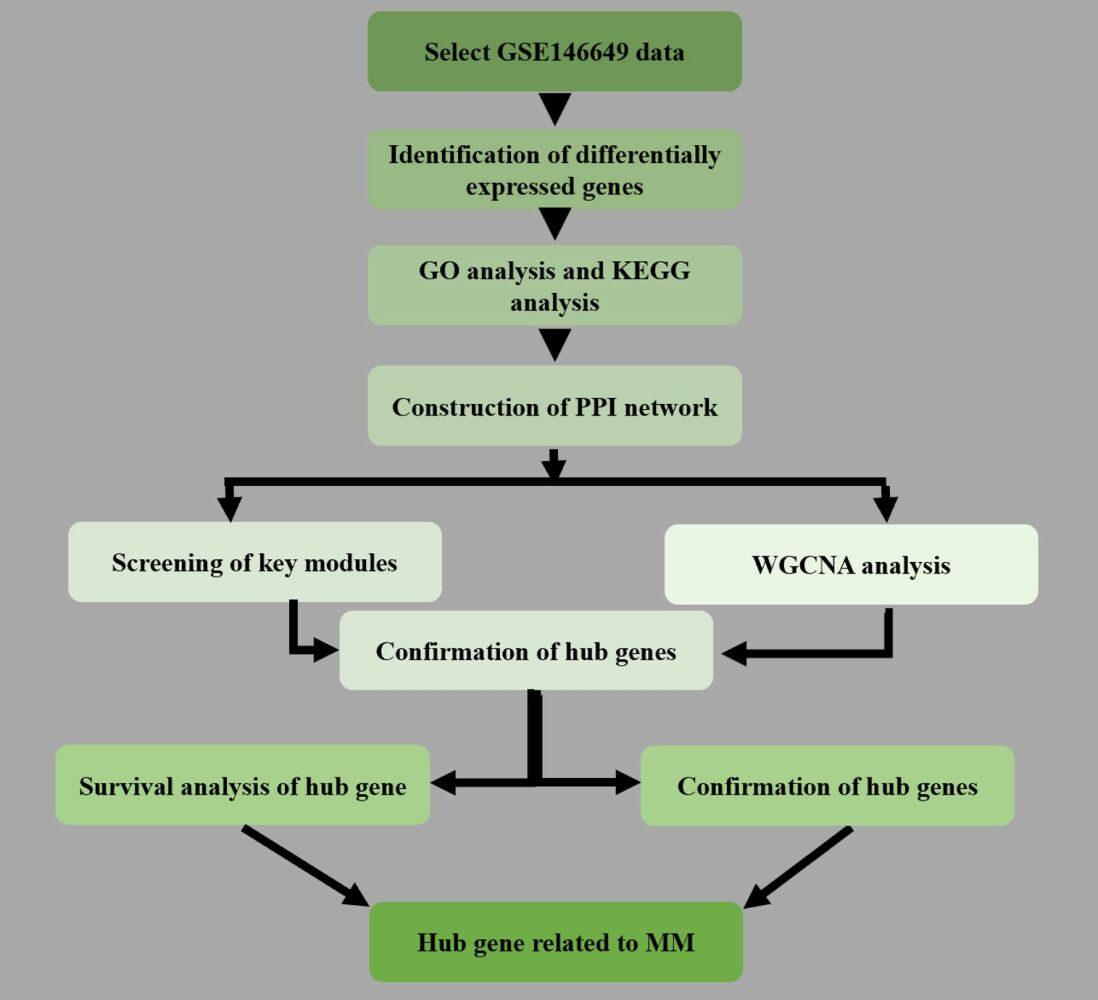
Objective: To study the differentially expressed genes between multiple myeloma and healthy whole blood samples by bioinformatics analysis, find out the key genes involved in the occurrence, development and prognosis of multiple myeloma, and analyze and predict their functions.
Methods: The gene chip data GSE146649 was downloaded from the GEO expression database. The gene chip data GSE146649 was analyzed by R language to obtain the genes with different expression in multiple myeloma and healthy samples, and the cluster analysis heat map was constructed. At the same time, the protein-protein interaction (PPI) networks of these DEGs were established by STRING and Cytoscape software. The gene co-expression module was constructed by weighted correlation network analysis (WGCNA). The hub genes were identified from key gene and central gene. TCGA database was used to analyze the expression of differentially expressed genes in patients with multiple myeloma. Finally, the expression level of TNFSF11 in whole blood samples from patients with multiple myeloma was analyzed by RT qPCR.
Results: We identified four genes (TNFSF11, FGF2, SGMS2, IGFBP7) as hub genes of multiple myeloma. Then, TCGA database was used to analyze the survival of TNFSF11, FGF2, SGMS2 and IGFBP7 in patients with multiple myeloma. Finally, the expression level of TNFSF11 in whole blood samples from patients with multiple myeloma was analyzed by RT qPCR.
Conclusion: The study suggests that TNFSF11, FGF2, SGMS2 and IGFBP7 are important research targets to explore the pathogenesis, diagnosis and treatment of multiple myeloma.
Keywords: multiple myeloma, bioinformatics, RT qPCR, PPI
Graphical Abstract:
Introduction
Multiple myeloma is a B-cell malignancy characterized by the malignant proliferation of clonal plasma cells in bone marrow,1 It is the second most common hematological malignancy after lymphoma.2 The age-standardised rate (ASR) of multiple myeloma incidence was 1·78 (95% UI 1·69-1·87) per 100 000 people globally and mortality was 1·14 (95% UI 1·07-1·21) per 100 000 people globally in 2020.3 Although the treatment strategies for multiple myeloma,4,5 such as proteasome inhibitors, immunomodulatory drugs, autologous stem cell transplantation and cellular immunotherapy, continue to improve, due to the characteristics of drug resistance and frequent recurrence of multiple myeloma, multiple myeloma is still incurable and the 5-year survival rate is approximately 50%6,7. At present, there is no clinical treatment plan to cure multiple myeloma, mainly through early chemotherapy and other methods to control the disease, but the treatment effect is not ideal. Studies have shown that 80–90% of patients with multiple myeloma have been troubled by active bone diseases, such as bone pain, spontaneous fracture, hypercalcemia, etc.8 because the delicate balance between bone formation and bone destruction has been broken.9 This seriously affects the quality of life of myeloma patients and increases their mortality. Although some studies10,11 have found that there is a bidirectional cross talk between bone cells and immune cells. Immune cells participate in the physiological and pathological activities of bone,12 but the effects on bone of immune cells (such as macrophages, granulocytes and innate immune pathways) remain unclear.13 Therefore, it is urgent to implement effective prevention, treatment and prognosis monitoring for patients with multiple myeloma. With the development of the biological big data era, it is possible to analyze biological information containing a large amount of data through the combination of biology, computer science and information technology. High throughput microarray is widely used in molecular diagnosis, tumor classification, prognosis and predictive analysis of tumors.14 It is expected to provide a new way to study the pathogenesis and molecular diagnosis of multiple myeloma (MM). In this study, bioinformatics technology was used to analyze the gene expression microarray data of patients with multiple myeloma (MM) and healthy control group, mine the differentially expressed genes of patients with multiple myeloma (MM) and healthy control group, screen the key genes through differential expression gene analysis, functional enrichment and survival analysis, and explore the changes of gene pathways and functions related to multiple myeloma (MM), The aim is to provide new research ideas and theoretical basis for the early diagnosis and gene therapy of multiple myeloma (MM).
Methods and Materials
Microarray Data
In order to explore the important genes related to the occurrence and development of multiple myeloma, the original gene expression profile of GSE146649 data set was obtained from the comprehensive gene expression database (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The mRNA expression profile of GSE146649 data set was detected using GPL570 platform (Affymetrix human genome U133 Plus 2.0 array [hg-u133_plus_2] Affymetrix human genome U133 Plus 2.0 array). In the study, 40 samples from the GSE146649 data set were selected for analysis, including samples initially diagnosed as multiple myeloma, samples of recurrent multiple myeloma, samples of fully remitted multiple myeloma and samples of normal people.
Identification of Differentially Expressed Genes
The original data set of GSE146649 is analyzed by using GEO2R software (https://www.ncbi.nlm.nih.gov/geo/geo2r/), a network tool using R’s “limma” software package to screen DEGs between multiple myeloma and normal tissues. Gene annotation and corresponding data files of DEGs were extracted by R software. The threshold value was |log2foldchange| >1.0 and adj.P.Val <0.05.
GO Analysis and KEGG Analysis
In order to further systematically analyze the functional annotation of differentially expressed genes (DEGs), go enrichment analysis and KEGG pathway analysis of differentially expressed genes (DEGs) were performed with David, an online biological information annotation database. The q value <0.05 was used as the cutoff value.
Construction of Protein-Protein Interaction Network
In order to explore the interaction between DEGs, the protein interaction (PPI) network of multiple myeloma differentially expressed genes (DEGs) was constructed by using string15 and Cytoscape online analysis tools, so as to find the hub gene.
Screening of Key Modules
The data set obtained from string analysis is imported into Cytoscape, and Cytohubba, a plug-in of Cytoscape,16 is used to select the central gene of biological network. In this study, the first 15 genes were selected as the central genes.
Coexpression and Module Function Analysis
Weighted gene co-expression network analysis (WGCNA) is a biological network that describes the correlation between differentially expressed genes (DEG).17 Co-expression analysis is a multidirectional network in which each node represents a gene. After evaluating the expression profile, the co-expression network was constructed by using the DEGs of GSE146649 dataset. The co-expression network of DEGs was constructed by R-package WGCNA. In addition, the Pearson correlation coefficient of genes was calculated to obtain the similarity matrix. In order to make the network conform to the scale-free network distribution, the appropriate weight value is selected and calculated. Select the appropriate soft threshold to measure the connectivity between genes. The adjacency matrix is transformed into a topological overlap matrix (TOM); At the same time, WGCNA package is used to cluster the matrix. For the generated cluster tree, the dynamic tree cutting method is used to cut the gene cluster tree. Genes with similar expression patterns were assigned to a branch, and each branch represented a co-expression module. In this study, the soft threshold is 6 and the minimum module size is set to 30. After analyzing the correlation between modules and clinical features, we selected the hub module which is most relevant to multiple myeloma. Gene significance (GS) represents the association between genes and clinical features. Module members (mm) reveal the relationship between module characteristic genes and gene expression.
Venn Diagram of Hub Genes
The crossing of central genes obtained by Cytohubba and key network modules constructed by WCGNA is considered to be a real hub gene, which is more related to the occurrence and development of multiple myeloma (MM). In order to achieve this intersection, the Lian chuan biological cloud platform (https://www.omicstudio.cn/tool) draw a Venn diagram.
Survival Analysis of Hub Genes
Then, the RNA sequencing data and matched clinical characteristics of multiple myeloma obtained from the Cancer Genome Atlas (TCGA) were used to analyze the impact of different expressions of hub genes on the survival of patients with multiple myeloma (MM).
Verification and Analysis of Hub Genes
Patient and Tissue Samples
(P)–Patient: From October 2020 to October 2021, 19 newly diagnosed multiple myeloma (MM) patients were selected from the Department of Hematology, Zhongshan Hospital Affiliated to Xiamen University, including 7 males and 12 females, with a median age of 61 (42 ~ 71) years. All the selected patients were diagnosed by clinical, myelogram, immunofixation electrophoresis, M protein quantification, whole-body flat bone radiography, bone scan, CT, MRI and other examinations. The diagnosis and staging were based on the Chinese guidelines for the diagnosis and treatment of multiple myeloma.18
(I)– Intervention: All patients in our hospital were treated with chemotherapy based on lenalidomide, bortezomib and dexamethasone.
(C)– Comparison: 10 healthy donors were selected as control group one. At the same time, 19 newly diagnosed multiple myeloma (MM) patients served as control group two after three months of standardized treatment.
(O)– Outcome: Compare the changes of RNA expression level (RT qPCR).
(T)–Time: The outcome would be measured after standardized treatment for three months.
In this study, 3mL heparin anticoagulation peripheral blood samples were collected before and after treatment. Cells were extracted by centrifugation within 24h and frozen at −80 °C. In this study, the use of patient organization and the acquisition of patient information were approved by the ethics committee of Zhongshan Hospital Affiliated to Xiamen University.(We ensured that all subjects obtained informed consent before the experiment was enrolled. The study was conducted in accordance with the principles of the declaration of Helsinki.)
RT qPCR Detection of RNA Expression Level
RNA extraction and quantitative polymerase chain reaction. RNA isolation of fresh multiple myeloma tissue samples was performed with Trizol reagent (Roche). Complementary DNA (cDNA) was synthesized by reverse transcription using hifiscript Kit (Invitrogen). Quantitative polymerase chain reaction was performed on TNFSF11 (RANKL) and GAPDH using SYBR Green qPCR master kit (Roche). GAPDH was selected as the internal reference gene. The primer sequence is as follows: STMN2 forward 5- tctcttgctgttgctgatgg-3; STMN2 reverse 5-ctcgtgagaccttcgctctt-3; GAPDH forward 5-CAAGGTCATCCATGACAACTTTG-3; GAPDH reverse 5- GTCCACCACCCTGTTGCTGTAG-3.
Results
Identification and Enrichment of DEGs in Multiple Myeloma
The GEO2R tool is used to analyze the DEGs in GSE146649. We screened the 424 DEGs between multiple myeloma and normal tissues, and found 350 up-regulated genes and 74 down regulated genes. DEGs are displayed in the volcano map and heat map (Figures 1 and 2). In GO analysis (Figure 3), biological process (BP) is mainly enriched in transcription regulation, cell adhesion and cell differentiation of RNA polymerase II promoter. Cellular components (CC) are mainly extracellular exosomes, cytoplasm, cytoplasmic membrane, etc. Molecular function (MF) is significantly enriched in protein binding rate. Further analysis of KEGG pathway enrichment of differentially expressed genes showed that the differentially expressed genes were mainly enriched in PI3K Akt signaling pathway, actin cytoskeleton regulation, AGE-RAGE signaling pathway in diabetes complications, HIF-1 signaling pathway, TGF-β Signaling pathway, Rap1 signaling pathway, and tumor necrosis factor signaling pathway. (Figure 4).
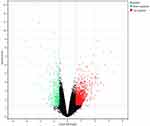 |
Figure 1 Volcano map of DEGs between MM case group and normal control group. The red plots in the volcano represent up-regulation and the green points represent down-regulation. |
 |
Figure 3 GO term enrichment analysis of module genes. |
Selection of Key Modules
Through string analysis, 424 DEGs are input into the PPI network, including 302 nodes and 1165 edges (Figure 5). Cytohubba is a plug-in that ranks nodes according to their network capabilities.16 It provides 11 analysis methods, including maximum neighborhood component density, maximum Group Centrality, maximum neighborhood component, degree, edge penetration component and six centralities (bottleneck, heart rate deviation, proximity, radius, medium and stress). This study used all 11 analysis methods in the PPI network to identify the top 15 genes (Figure 6). According to 11 analysis methods of Cytohubba, 48 genes were screened. This finding may indicate that 48 genes play an important role in multiple myeloma.
 |
Figure 5 424 DEGs were filtered into the DEGs PPI network complex that contained 302 nodes and 1165 sides. |
Construction and Analysis of the Key Gene Co-Expression Network
A total of 40 clinical samples of GSE146649 were collected for analysis. WGCNA performs R language analysis and divides highly related genes into one module choice β= 6 (scale-free r2=0.90) as the appropriate soft threshold to ensure scale-free analysis. Six modules were identified. It was found that the blue module was mainly related to multiple myeloma (Figures 7 and 8). In addition, the relationship between modules and clinical features was also studied for gene significance (GS) and module members (MM) in the six modules. The heat map of the phenotypic correlation between modules and clinical features is shown in the figure (Figure 9). We can see that the blue module has the most significant correlation with multiple myeloma (diagnosis), which is 0.94, and the p value is statistically significant. Later, the scatter diagram of the correlation between gene significance (GS) and module members (MM) (Figures 10 and 11) was carried out. We can see that the blue module has the most significant correlation with multiple myeloma (diagnosis), while the blue module has the least significant correlation with health. Finally, according to the critical values: module member value >0.80 and GS value >0.2, the gene in the blue module is selected as the central gene. A total of 295 central genes from the blue module were selected for further analysis.
 |
Figure 8 Heatmap plot of the adjacencies of modules. Red represents high adjacency (positive correlation) and blue represents low adjacency (negative correlation). |
 |
Figure 10 Scatter diagram of blue module and initially diagnosis. |
 |
Figure 11 Scatter diagram of blue module and healthy. |
Identification of Key Genes
295 genes constructed by WGCNA were selected as central genes. In the PPI network, 48 genes identified by Cytohubba were defined as hub genes. Finally, the intersection of Cytohubba and WGCNA is four genes (TNFSF11, FGF2, SGMS2, IGFBP7), which are considered the “real” hub genes (Figure 12).
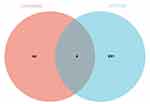 |
Figure 12 Venn diagram of hub genes. |
Survival Analysis of Hub Gene
Next, the survival condition of four hub genes were analyzed. Survival is affected by many factors, including treatment strategies, patients’ physical status, tumor characteristics and so on. The markers related to these factors are often the key to the prognosis evaluation of patients with multiple myeloma. Targeted therapy against these markers will become a breakthrough in the treatment of multiple myeloma in the future. In this study, the TCGA database was used to analyze the relationship between key genes and prognosis. It was found (Figure 13) that the long-term overall survival rate of multiple myeloma with TNFSF11, FGF2, SGMS2 and IGFBP7 mutations was significantly lower, and the difference was statistically significant; This indicates that the differential expression of target genes has adverse effects on the survival and overall survival of patients with multiple myeloma. Therefore, the screening of four target genes (TNFSF11, FGF2, SGMS2, IGFBP7) is the key molecule for the occurrence and development of multiple myeloma, and can be used as an important target for the diagnosis and treatment of multiple myeloma.
Verification of Key Genes
qPCR Detection
Multiple myeloma whole blood samples and normal whole blood samples were evaluated by RT-qPCR to verify whether TNFSF11 was differentially expressed in the samples.
The expression of TNFSF11 and internal reference GAPDH in 29 samples was detected by qPCR. The gene amplification curve and dissolution curve are shown in Figures 14 and 15. The expression level of TNFSF11 in whole blood samples of patients with multiple myeloma was significantly higher than that of healthy donors (Figure 16). After treatment, the expression level of TNFSF11 in whole blood samples of patients with multiple myeloma decreased (Figure 17).
 |
Figure 14 TNFSF11 (RANKL) amplification curve. |
 |
Figure 15 TNFSF11 (RANKL) dissolution curve. |
Discussion
Multiple myeloma is the second most common hematological malignancy after lymphoma. There is no clinical treatment plan for multiple myeloma. The disease is mainly controlled by early chemotherapy, but the treatment effect is not ideal. Therefore, it is very important to screen early diagnostic markers and regulatory pathways related to multiple myeloma. In this study, we used the gene expression profile of GSE146649 from 31 patients with multiple myeloma and 9 normal subjects to find some biomarkers and clarify the molecular mechanism of multiple myeloma.
The study found that 424 DEGs were associated with multiple myeloma, including 350 up-regulated genes and 74 down regulated genes. PPI and WGCNA analyses were used to select central genes associated with multiple myeloma. In addition, the related functions and pathways of multiple myeloma were analyzed. In GO analysis, DEG is mainly enriched in the transcription regulation, cell adhesion, cell differentiation of RNA polymerase II promoter, significant enrichment in protein binding rate, cell adhesion is the key to the spread and progress of multiple myeloma. Research shows that MM plasma cell trafficking and homing to the BM are regulated by soluble factors, mainly chemokines such as CXCL12, as well as by direct cell–cell interactions through adhesion molecules expressed by plasma and bone marrow-resident cells.19 The final phases in MM involve the egress of myeloma cells from the BM to the bloodstream to colonize different organs, once they become independent from growth signals provided by the BM milieu, a condition named extramedullary disease.20,21 Extramedullary multiple myeloma remains challenging from a therapeutic and biological perspective.22
Further analysis of KEGG pathway enrichment of differentially expressed genes showed that the differentially expressed genes were mainly enriched in PI3K Akt signaling pathway, actin cytoskeleton regulation, AGE-RAGE signaling pathway in diabetes complications, HIF-1 signaling pathway and TGF-β Signaling pathway, Rap1 signaling pathway, and tumor necrosis factor signaling pathway. We know that the ability of bone formation depends on the activity of osteoblasts and the osteogenic differentiation ability of bone marrow stromal cells (BMSCs). Therefore, in order to fully understand the pathogenesis of multiple myeloma and explore its characteristics, it is feasible to study from two aspects. Pi3k/akt signaling pathway is the key signaling pathway for osteogenic differentiation of bone marrow mesenchymal stem cells.23 It can promote chondrocyte proliferation and inhibit autophagy. Relevant studies have confirmed that pi3k/akt signaling pathway is involved in pathological bone diseases such as osteoporosis, osteoarthritis and osteosarcoma,24,25 and regulates the proliferation, differentiation and apoptosis of osteoclasts and osteoblasts.26,27 It is reported28 that AGEs modified collagen can promote the release of intracellular reactive oxygen species, interfere with the adhesion between osteoblasts and matrix, and inhibit the differentiation and proliferation of osteoblasts. Rage and its ligands play an important physiological role in bone. The increase of rage and its ligands is obviously related to various bone related diseases.29 HIF-1 signaling pathway plays an important role in bone formation and bone resorption, and is closely related to the promotion of osteoblast bone formation, this signaling pathway can increase the number of osteoclasts and osteoclastic activity.30 TGF- β signaling pathway can make progenitor cells enter adipogenic or osteoblastic lineage by recombining cytoskeleton, so as to improve the osteogenic ability of mesenchymal stem cells.31
WGCNA analysis showed that the six modules had related expression patterns. 296 hub genes related to metastasis were selected from the blue module. Based on the plug-in Cytohubba in Cytoscape, four key genes were screened. TNFSF11, FGF2, SGMS2 and IGFBP7. FGF2 is a multifunctional growth factor belonging to the FGF family. FGF2 is an efficient mitogen, which can promote the proliferation of osteoblast precursor cells.32 In addition, FGF2 can inhibit the aging of BMSCs and promote cell proliferation through pi3k/akt-mdm2 pathway, to maintain the stem cell characteristics of BMSCs.33 Sgms2 is a sphingomyelin synthetase SMS2 that is significantly expressed in cortical bone and encodes on the plasma membrane. Some research results show34 that SMS2 has bone regulatory properties. The mutation of SGMS2 is the basis of a series of bone diseases, including simple osteoporosis and complex bone dysplasia, indicating that plasma membrane combined with sphingomyelin metabolism plays a key role in the balance of the bone environment. IGFBP7 belongs to the insulin-like growth factor binding proteins (IGFBPs) family, which consists of six high affinity and several low affinity members,35 and plays an important role in bone metabolism. It has been reported36 that the role of IGFBP7 involves insulin resistance, which is considered to be closely related to the development of osteoporosis. Ye et al. Found37 that IGFBP7 inhibits osteoclast formation in vitro and in vivo, suggesting that IGFBP7 is a negative regulator of osteoclast formation and plays a protective role in osteoporosis. Zhang et al. Found38 that IGFBP7 increased OPG expression in vivo and enhanced osteogenic ability. We know osteoprotegerin (OPG) and nuclear factor κ B receptor activating factor (RANK) are members of the tumor necrosis factor-α receptor superfamily,39 TNFSF11 (RANKL) is a transmembrane receptor, belonging to the tumor necrosis factor (TNF) superfamily. TNFSF11 (RANKL) is mainly distributed on the surface of osteoblasts and bone marrow stromal cells, and plays an important role in bone formation.40
The combination of TNFSF11 (RANKL) and RANK can promote the differentiation of osteoclasts, while OPG can inhibit the differentiation and activity of osteoclasts,41 delaying the activation of osteoclast precursor cells and maintain the balance of bone metabolism. Therefore, RANK, RANKL (TNFSF11) and OPG are the key regulatory mechanisms to control the differentiation and activity of osteoblasts and osteoclasts.42 The ratio of RANKL (TNFSF11)/OPG is an important lever index to evaluate bone health and regulate the balance of bone resorption.43 This study found that the expression of TNFSF11 (RANKL) in multiple myeloma was up-regulated compared with the normal group. TNFSF11 is highly expressed in whole blood samples of patients with multiple myeloma. The expression level of TNFSF11 (RANKL) in peripheral blood of patients with multiple myeloma is closely related to osteolytic osteopathy, which can reflect the short-term bone metabolism.44
In this study, TNFSF11, FGF2, SGMS2 and IGFBP7 were selected as important key genes in the pathogenesis of mm by bioinformatics method, and then the levels of TNFSF11 (RANKL) in peripheral blood of patients with multiple myeloma diagnosed in our hospital before and after treatment were analyzed and compared by PCR technology. We found that the expression level of TNFSF11 (RANKL) in peripheral blood of patients with multiple myeloma decreased significantly after treatment, and compared with healthy patients, TNFSF11 (RANKL) was highly expressed in patients with multiple myeloma, suggesting that TNFSF11 (RANKL) may play a pathogenic role in multiple myeloma. In addition, in this study, we used comprehensive analysis from WGCNA and DEGs to screen potential biomarkers, which may also make our results more credible. The biological functions and potential molecular mechanisms of the key genes TNFSF11, FGF2, SGMS2 and IGFBP7 explored in this study still need to be explored in future research.
Conclusion
In conclusion, through comprehensive bioinformatics analysis, TNFSF11, FGF2, SGMS2 and IGFBP7 were screened as key genes related to multiple myeloma, and their roles and pathways in multiple myeloma were discussed. At the same time, RT-qPCR was used to preliminarily confirm that TNFSF11 was highly expressed in whole blood samples of patients with multiple myeloma. Combined with the clinical conditions of patients and current relevant research progress, we believe that the above four key genes are still worth further exploration and analysis. The results of this study increase our understanding of biomarkers and potential therapeutic targets of multiple myeloma. However, we need to further study the role and mechanism of TNFSF11, FGF2, SGMS2 and IGFBP7 in the future development of multiple myeloma.
Patient and Tissue Samples
All samples were from the Department of Hematology, Zhongshan Hospital, Xiamen University.
Ethics Approval and Consent to Participate
This study was conducted with approval from the Ethics Committee of Zhongshan Hospital Affiliated to Xiamen University. GEO belongs to public databases. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data and therefore has no other conflicts of interest.
Acknowledgments
All authors would like to thank the patients and their families for their contribution to this study. All authors would like to thank the GEO database.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by Natural Science Foundation of Fujian Province (Grant Code: 2019D009).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cook G, Orcid I, Morris CT. Evolution or revolution in multiple myeloma therapy and the role of the UK. Br J Haematol. 2020;191:542–551. doi:10.1111/bjh.17148
2. Chen J, Garssen J, Redegeld F. The efficacy of bortezomib in human multiple myeloma cells is enhanced by combination with omega-3 fatty acids DHA and EPA: timing is essential. Clin Nutr. 2021;40:1942–1953.
3. Huang J, Chan SC, Lok V, et al. The epidemiological landscape of multiple myeloma: a global cancer registry estimate of disease burden, risk factors, and temporal trends. Lancet Haematol. 2022. doi:10.1016/S2352-3026(22)00165-X
4. Mikkilineni L, Kochenderfer JN, Orcid I. CAR T cell therapies for patients with multiple myeloma. Nat Rev Clin Oncol. 2021;18:71–84. doi:10.1038/s41571-020-0427-6
5. Sonneveld P, Broijl A. Treatment of relapsed and refractory multiple myeloma. Haematologica. 2016;101:396–406. doi:10.3324/haematol.2015.129189
6. Goodman AM, Kim MS, Prasad V. Persistent challenges with treating multiple myeloma early. Blood. 2021;137:456–458. doi:10.1182/blood.2020009752
7. Tabata M, Tsubaki M, Takeda T, et al. Inhibition of HSP90 overcomes melphalan resistance through downregulation of Src in multiple myeloma cells. Clin Exp Med. 2020;20:63–71.
8. Terpos E, Dimopoulos MA, Ntanasis-Stathopoulos I, et al. Myeloma bone disease: from biology findings to treatment approaches. Blood. 2019;33(14):1534–1539. doi:10.1182/blood-2018-11-852459
9. Zarghooni K, Hopf S, Eysel P. Management of osseous complications in multiple myeloma. Der Internist. 2019;60:42–48. doi:10.1007/s00108-018-0530-2
10. Nam D, Mau E, Wang Y, et al. T-lymphocytes enable osteoblast maturation via IL-17F during the early phase of fracture repair. PLoS One. 2012;7:e40044. doi:10.1371/journal.pone.0040044
11. Mori G, D’ Amelio P, Faccio R, Brunetti G. The Interplay between the bone and the immune system. Clin Dev Immunol. 2013;2013:720504.
12. Arron JR, Choi Y. Bone versus immune system. Nature. 2000;408:535–536. doi:10.1038/35046196
13. Gnoni A, Brunetti O, Longo V, et al. Immune system and bone microenvironment: rationale for targeted cancer therapies. Oncotarget. 2020;11:480–487. doi:10.18632/oncotarget.27439
14. Yin L, Cai Z, Zhu B, Xu C. Identification of key pathways and genes in the dynamic progression of HCC based. Genes. 2018;9:92. doi:10.3390/genes9020092
15. von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi:10.1093/nar/gkg034
16. Chin CH, Chen SH, Wu HH, et al. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8:S11.
17. Zhang J, Lan Q, Lin J. Identification of key gene modules for human osteosarcoma by co-expression. World J Surg Oncol. 2018;16:89.
18. Chen WM. The guidelines for the diagnosis and management of multiple myeloma in China. Zhonghua nei ke za zhi. 2017;56:799–800. doi:10.3760/cma.j.issn.0578-1426.2017.11.004
19. García-Ortiz A, Orcid I, Rodríguez-García Y, et al. The role of tumor microenvironment in multiple myeloma development. Cancers. 2021;13:217. doi:10.3390/cancers13020217
20. Ghobrial IM. Myeloma as a model for the process of metastasis: implications for therapy. Blood. 2012;120(1):20–30. doi:10.1182/blood-2012-01-379024
21. Da Vià MC, Orcid I, Solimando AG, et al. CIC mutation as a molecular mechanism of acquired resistance to combined BRAF-MEK. Oncologist. 2020;25:112–118. doi:10.1634/theoncologist.2019-0356
22. Radhika B, Sagar R, Kumar S. Extramedullary disease in multiple myeloma. Blood Cancer J. 2021;11:161.
23. Huang X, Huang D, Zhu T, et al. Sustained zinc release in cooperation with CaP scaffold promoted bone regeneration via directing stem cell fate and triggering a pro-healing immune stimuli. J Nanobiotechnology. 2021;19:20.
24. Lin C, Shao Y, Zeng C, et al. Blocking PI3K/AKT signaling inhibits bone sclerosis in subchondral bone and attenuates post‐traumatic osteoarthritis. J Cell Physiol. 2018;233:6135–6147. doi:10.1002/jcp.26460
25. Xi JC, Zang HY, Guo LX, et al. The PI3K/AKT cell signaling pathway is involved in regulation of osteoporosis. J Recept Signal Transd Res. 2015;35:640–645. doi:10.3109/10799893.2015.1041647
26. Gu YX, Du J, Si MS, et al. The roles of PI3K/Akt signaling pathway in regulating MC3T3-E1 preosteoblast. J Biomed Mater Res A. 2013;101:748–754. doi:10.1002/jbm.a.34377
27. Zou W, Yang S, Zhang T, et al. Hypoxia enhances glucocorticoid-induced apoptosis and cell cycle arrest via the PI3K/Akt signaling pathway in osteoblastic cells. J Bone Miner Metab. 2015;33(6):615–624. doi:10.1007/s00774-014-0627-1
28. Yamagishi S, Fukami K, Matsui T. Crosstalk between advanced glycation end products (AGEs)-receptor RAGE axis and dipeptidyl peptidase-4-incretin system in diabetic vascular complications. Cardiovasc Diabetol. 2015;14:1–2
29. Plotkin LI, Essex AL, Davis HM. RAGE signaling in skeletal biology. Curr Osteoporos Rep. 2019;17:16–25. doi:10.1007/s11914-019-00499-w
30. Knowles HJ, Cleton-Jansen AM, Korsching E, Athanasou NA. Hypoxia-inducible factor regulates osteoclast-mediated bone resorption. FASEB J. 2010;24:4648–4659. doi:10.1096/fj.10-162230
31. Elsafadi M, Manikandan M, Almalki S, et al. TGFβ1-induced differentiation of human bone marrow-derived MSCs is mediated by changes to the actin cytoskeleton. Stem Cells Int. 2018;2018:6913594. doi:10.1155/2018/6913594
32. Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463–1486. doi:10.1101/gad.266551.115
33. Coutu DL, François M, Galipeau J. Inhibition of cellular senescence by developmentally regulated FGF receptors in mesenchymal stem cells. Blood. 2011;117(25):6801–6812. doi:10.1182/blood-2010-12-321539
34. Pekkinen M, Terhal PA, Botto LD, et al. Osteoporosis and skeletal dysplasia caused by pathogenic variants in SGMS2. JCI Insight. 2019;4. doi:10.1172/jci.insight.126180
35. Mohan S, Baylink DJ. IGF-binding proteins are multifunctional and act via IGF-dependent and IGF-Independent Mechanisms. Am Zool. 2001;41:19–31.
36. Liu Y, Wu M, Ling J, et al. Serum IGFBP7 levels associate with insulin resistance and the risk of metabolic syndrome in a Chinese population. Sci Rep. 2015;5:10227.
37. Ye C, Hou W, Chen M, et al. IGFBP7 acts as a negative regulator of RANKL-induced osteoclastogenesis and oestrogen deficiency‐induced bone loss. Cell Prolif. 2020;53:e12752. doi:10.1111/cpr.12752
38. Zhang W, Chen E, Chen M, et al. IGFBP7 regulates the osteogenic differentiation of bone marrow-derived. FASEB J. 2018;32(4):2280–2291. doi:10.1096/fj.201700998RR
39. Fu D, Qin K, Yang S, et al. Proper mechanical stress promotes femoral head recovery from steroid-induced osteonecrosis in rats through the OPG/RANK/RANKL system. BMC Musculoskelet Disord. 2020;21:281.
40. Yasuda H, Id- Orcid X. Discovery of the RANKL/RANK/OPG system. BMC Musculoskelet Disord. 2020;21:2–11.
41. Yang B, Li S, Chen Z, et al. Amyloid β peptide promotes bone formation by regulating Wnt/β-catenin signaling. FASEB J. 2020;34(3):3583–3593. doi:10.1096/fj.201901550R
42. Lv WT, Du DH, Gao RJ, et al. Regulation of hedgehog signaling offers a novel perspective for bone homeostasis disorder treatment. Int J Mol Sci. 2019;20:3981
43. Chen B, Du Z, Dong X, et al. Association of Variant Interactions in RANK, RANKL, OPG, TRAF6, and NFATC1 Genes with the development of osteonecrosis of the femoral head. DNA Cell Biol. 2019;38:734–746.
44. Bolzoni M, Storti P, Bonomini S, et al. Immunomodulatory drugs lenalidomide and pomalidomide inhibit multiple. Exp Hematol. 2013;41:387–397.e381. doi:10.1016/j.exphem.2012.11.005
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








