Back to Journals » Therapeutics and Clinical Risk Management » Volume 17
Bioinformatics Analysis of a Functional ANGPT1 Variant That Interferes with miR-607 and Its Association with Susceptibility and Outcome of Ischemic Stroke in a Han Population
Authors Chen AN, Zhong LL, Ju K, Cao H
Received 16 July 2021
Accepted for publication 16 September 2021
Published 28 September 2021 Volume 2021:17 Pages 1045—1052
DOI https://doi.org/10.2147/TCRM.S328964
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Deyun Wang
Ai-Nian Chen, Ling-Ling Zhong, Ke-ju Ju, Hua Cao
Department of Neurology, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, Huaian, Jiangsu, People’s Republic of China
Correspondence: Ai-Nian Chen
Department of Neurology, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, 1 Huanghe Road West, Huaian, Jiangsu, 223300, People’s Republic of China
Tel +86 18751292677
Email [email protected]
Purpose: Ischemic stroke (IS) is a major cause of disability and death. We used bioinformatics approaches to investigate a functional ANGPT1 variant that interferes with miR-607 and explored its association with IS.
Materials and Methods: An IS expression microarray (GSE16561) was downloaded from the GEO and used to identify differentially expressed genes (DEGs) and functional enrichment pathways. Analyses showed that ANGPT1 participated in six key pathways and was susceptible to a key functional polymorphism rs2507799. We genotyped 567 IS patients and 500 controls for ANGPT1 rs2507799. Luciferase assays were also conducted to investigate the binding between miR-607 and ANGPT1 rs2507799.
Results: In total, we identified 458 DEGs between IS patients and healthy controls in the GSE16561 dataset. GO functional enrichment analysis showed that these DEGs were mainly enriched in cell-substrate junctions, the regulation of peptide secretion, and the regulation of cytokine secretion involved in immune response. ANGPT1 rs2507799 T-carriers had a significantly higher risk of IS (Dominant model: OR = 1.48, 95% CI = 1.01– 2.17, P = 0.044). IS patients harboring the TC/TT genotype experienced significantly more severe injuries in terms of neurological function (Dominant model: OR = 2.06, 95% CI = 1.28– 3.31, P = 0.003). Analysis also showed that IS patients harboring the TC/TT genotype had a significantly worse outcome (Dominant model: OR = 2.22, 95% CI = 1.35– 3.67, P = 0.002). Luciferase assays indicated that miR-607 could affect luciferase activity by binding to the ANGPT1 mutant type.
Conclusion: In this study, we used bioinformatical methods to investigate a key IS-related gene ANGPT1 and its functional polymorphism rs2507799. rs2507799 was found to be associated with a significantly increased risk for IS, a significantly more severe initial stroke severity, and a worse outcome. These results may help to improve the future management of ischemic stroke.
Keywords: ischemic stroke, functional enrichment analysis, polymorphism, ANGPT1, miRNAs
Introduction
Ischemic stroke (IS) is caused by the sudden loss of blood circulation to an area of the brain that causes injury to neurological function and represents a major cause of global disability and mortality.1 IS is known to be a heterogeneous and multifactorial disease. Genetic factors, particularly those involving environmental interactions, are known to play important roles in IS susceptibility and outcomes.2
Genetic variations in the 3ʹ-untranslated region (3ʹ-UTR) of mRNA have been reported to be associated with a variety of diseases.3–5 One potential theory to explain these associations is the post-transcriptional regulation of gene expression by microRNAs (miRNAs).6 The basic concept underlying this mechanism is that the affinity of miRNA to mRNA can be changed by mutation in the 3ʹ-UTR of mRNA, thus affecting mRNA degradation and final gene expression at the post-transcriptional level.
In this study, we hypothesized that genetic variations in the key molecular pathways associated with IS may exert influence on susceptibility and outcome. We used bioinformatical methods to investigate the key molecular pathways involved in IS. The Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) is a database that is commonly used for bioinformatics analysis. We downloaded a matrix file (GSE16561) from the GEO that contained IS expression profile data. Then, we identified differentially expressed genes (DEGs) and performed functional enrichment analysis. We found that angiopoietin 1 (ANGPT1) was significantly enriched in IS and participated in six key functional pathways. ANGPT1 is known to be involved in angiogenesis and the anti-inflammatory process; these processes play key roles in neural protection after stroke. Taking these factors into consideration, we hypothesized that genetic variability within this gene may play an important role in the occurrence and outcome of IS. Next, we used bioinformatics software to identify and characterize candidate single nucleotide polymorphisms (SNPs) that might influence the expression of ANGPT1 by miRNA analysis. We successfully identified ANGPT1 rs2507799 as a potential SNP and investigated the potential relationships between this SNP and IS in terms of susceptibility and outcomes by comparing between IS patients and healthy controls recruited from our hospital. Finally, we used luciferase assays to confirm the regulatory ability of ANGPT1 rs2507799 on IS.
Materials and Methods
Study Subjects
In this study, we recruited 567 cases of IS from The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University between June 2017 and December 2019. We also recruited 500 controls without IS. The diagnosis of ischemic stroke was made in accordance with the criteria put forward by the World Health Organization.7 All patients and controls completed an epidemiological questionnaire featuring demographical information, medical history (such as hypertension and diabetes mellitus), and other vascular risk factors. This study was conducted in accordance with the Declaration of Helsinki. All patients signed an informed consent form and the study was approved by the Ethics Committee of the Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University (Project identification code: KY-P-2017-012-01).
Stroke Severity and Functional Outcome Assessments
Upon hospital admission, the National Institutes of Health Stroke Scale (NIHSS) was used to assess stroke severity. Because the NIHSS scores were skewed and did not comply with a normal distribution, the data were dichotomized for logistic regression analysis. In accordance with previous studies, the cutoff value for mild and severe stroke was set at an NIHSS score of 6.8,9 One month after the patients were discharged, we used the modified Rankin Scale (mRS) to evaluate disability in each IS patient, as previously described.10,11 In accordance with previous studies, an mRS score of 0–2 was defined as a better functional outcome, and an mRS score of 3–6 was defined as a worse functional outcome.12
Identification and Functional Enrichment Analysis of DEGs
The Affy13 package and limma package14 in R software were used to preprocess gene expression profile data and identify DEGs. In brief, data (in CEL format) were annotated and converted into expression measures. Next, we summarized the probe data from the GSE16561 dataset and acquired gene expression matrices. The t-test was used to identify DEGs with specific cut off criteria (|log [FC]| > 1.5 and a corrected P < 0.05). Next, Gene Ontology (GO) functional enrichment analysis was conducted using the clusterProfiler package.15 A false discovery rate (FDR) < 0.05 was set as the threshold value. Then, we recorded significant results for molecular function (MF), biological process (BP), and cellular component (CC).
SNP Selection and the Prediction of Binding Between miRNAs and SNPs
Next, we searched the GenBank Single Nucleotide Polymorphism (SNP) database (https://www.ncbi.nlm.nih.gov/snp) to identify potential ANGPT1 genetic variants in the 3ʹ-UTR using the following parameters: organism (Homo sapiens), function class (3ʹ-UTR), and Global MAF (0.05–0.4). Bioinformatics software (http://bioinfo.life.hust.edu.cn/miRNASNP/#!/) was then used to predict potential miRNAs that could be affected by SNPs.
DNA Extraction and Genotyping
Total DNA samples were extracted from patients and controls in accordance with a standard protocol.16 The TaqMan allelic discrimination assay was conducted to genotype samples for the SNP (Applied Biosystems, San Diego, CA, USA). The PCR conditions were as follows: 50°C for 2 mins, 95°C for 10 mins, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The allelic discrimination mode of SDS 2.3 software was used to calculate the results.
Blood Samples and Tests
Venous blood samples were collected from IS patients within 24 h of admission. Samples were centrifuged at 3000 rpm for 20 min and stored at −70°C within 30 min of collection. Plasma levels of ANGPT1 was measured with a commercial assay (R&D Systems, Minneapolis, MN, USA). The intra- and inter-assay coefficients of variation lay between 5% and 10%.
Plasmid Construction and the 3ʹ-UTR Luciferase Reporter Assay
The 3ʹ-UTR fragments containing ANGPT1 alleles (wild and mutant) were amplified by PCR. The PCR products were then cloned into the multiple cloning site of the pGL3-promoterless luciferase-based plasmid (Promega, CA, USA) between the BamHI and XhoI restriction sites (Genscript, Nanjing, China). For reporter assays, Lipofectamine 2000 (Invitrogen Corp, CA, USA) was used to transfect 293T cells with 100 ng of the pGL3-ANGPT1-wild construct, the pGL3-ANGPT1-mutant construct, or microRNA mimics. We also co-transfected cells with the Renilla luciferase vector pRL-SV40 (5 ng) to allow us to normalize for transfection efficiency.
Statistical Analysis
All statistical analyses were conducted using Stata/SE (V.12.0 for Windows; StataCorp LP, TX, USA). Demographic data were compared using two-sample t-tests and the Chi-squared (χ2) test. The relationships between genotypes and IS susceptibility, severity, and functional outcome, were estimated by odds ratios (ORs) and 95% confidence intervals (CIs) using multivariate logistic regression analysis. Luciferase activities and plasma ANGPT1 levels were compared between different genotype groups by independent sample t-tests. All statistical tests were two-sided and a P-value < 0.05 was considered as statistically significant.
Results
Demographic Characteristics of Patient and Control Groups
As shown in Table 1, there were no significant differences between the patient and control groups in terms of age or sex. Compared to the control group, the group of IS patients featured significantly higher proportions of patients with diabetes (P = 0.008), hypertension (P = 0.02), and drinking (P = 0.001). Patients in the IS group also exhibited higher levels of serum total cholesterol, triglycerides, and LDL-C.
 |
Table 1 Demographic Characteristics of IS Patients and Controls |
Identification and Functional Enrichment Analysis of DEGs
In the present study, we used the GSE16561 dataset to identify 458 DEGs between IS patients and healthy controls. Of these DEGs, 344 genes were upregulated and 114 genes were downregulated (Figure 1). GO functional enrichment analysis showed that these DEGs were mainly enriched in cell-substrate junction, cell-substrate adherens junction, the regulation of glial cell proliferation, the positive regulation of glial cell proliferation, the negative regulation of the production of molecular mediators of immune response, the regulation of peptide secretion, and the regulation of cytokine secretion involved in immune response (Figure 2 and Supplementary Table 1). Furthermore, we discovered that ANGPT1 participated in six of these key pathways. Thus, it is reasonable to assume that genetic variability within this gene may play an important role in the occurrence and outcome of IS.
The Selection of ANGPT1 SNPs and Genotype Frequencies in Patients and Controls
Searches of the NCBI SNP database identified two candidate ANGPT1 SNPs (Table 2). Next, we investigated the frequencies of these two SNPs in 567 cases of ischemic stroke and 500 controls. As shown in Table 3, there was a significantly different distribution of the ANGPT1 rs2507799 SNP when compared between the two groups. After adjustment for drinking, diabetes, hypertension, total cholesterol, triglycerides and LDL-C, logistic regression analysis indicated that subjects who had the mutant T allele had an increased risk for ischemic stroke (Dominant model: OR = 1.48, 95% CI = 1.01–2.17, P = 0.044).
 |
Table 2 SNPs Located in the ANGPT1 Gene 3ʹ-UTR and Candidate miRNAs Associated with ANGPT1 rs2507799 Predicted by Bioinformatics Analysis |
 |
Table 3 Genotype Frequencies of the ANGPT1 rs2507799 Polymorphism Among Patient Group and Control Group |
Relationships Between ANGPT1 rs2507799, Stroke Severity and Outcome
Next, we examined whether there were associations between the ANGPT1 rs2507799 polymorphism and the severity and outcome of IS. Logistic regression analysis indicated that IS patients who had the TC/TT genotype had a significantly more severe injury in terms of neurological function (Dominant model: OR = 2.06, 95% CI = 1.28–3.31, P = 0.003) (Table 4). Our analysis also revealed that IS patients with the TC/TT genotype had a significantly worse outcome in terms of disability (Dominant model: OR = 2.22, 95% CI = 1.35–3.67, P = 0.002) (Table 5).
 |
Table 4 Association of ANGPT1 rs2507799 Polymorphism with is Severity |
 |
Table 5 Association of ANGPT1 rs2507799 Polymorphism with is Outcome |
ANGPT1 rs2507799 Caused Changes in miRNA Binding
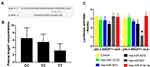
Because ANGPT1 rs2507799 was associated with the severity and outcome of IS, we next conducted bioinformatics analysis to predict if this SNP could affect the functional binding of miRNAs. Our analyses suggested that five miRNAs may be able to interact with the rs2507799 target site when the 3´-UTR of ANGPT1 is mutated (Table 1). Next, we transfected cells with the five candidate miRNAs mimics with either the wild or mutant form of ANGPT1. Luciferase assays indicated that miR-607 could lead to alterations in luciferase activity by binding to the ANGPT1 mutant type (Figure 3A and C). Analysis of the plasma levels of ANGPT1 also showed that patients carrying the TC/TT genotype had lower plasma concentrations of ANGPT1 than patients carrying the CC genotype (Figure 3B).
Discussion
To the best of our knowledge, this is the first study to use bioinformatic methods to identify and characterize ANGPT1 as a key IS-related gene and rs2507799 as a functional SNP of this critical gene. Our results indicated that ANGPT1 rs2507799 T-carriers had an increased risk for IS. We also found that IS patients carrying the rs2507799 T allele experienced a more severe initial injury in terms of neurological function, and a worse outcome. The potential mechanism underlying these observations, is that rs2507799 may alter the binding affinity of miR-607 to ANGPT1 mRNA, thus increasing the degradation of mRNA.
ANGPT1 is a ligand of endothelial-specific tyrosine kinase receptor (Tie2). When binding to ANGPT1, Tie2 can be phosphorylated; this leads to the activation of pathways involved in peri-endothelial cell recruitment, endothelial permeability, and anti-inflammatory signal transduction.17 Disruption of the blood–brain barrier (BBB), along with inflammatory processes, can lead to brain damage following ischemic stroke.18 Thus, the angiogenic and anti-inflammatory effects of ANGPT1 may be instrumental in neural protection after stroke. Previous animal studies reported that the upregulation of ANGPT1 prior to middle cerebral artery occlusion could reduce BBB leakage and cerebral ischemic volume in mice.19 Furthermore, the upregulation of ANGPT1 was also associated with an improvement in outcome in a rat model of middle cerebral artery occlusion.20 In addition, previous clinical study reported that a SNP of ANGPT1 was associated with the occurrence of stroke.18 In another clinical study, Golledge et al reported that the upregulation of plasma ANGPT1 levels was related to a better outcome after stroke;12 these previous findings were consistent with our present results.
A previous study reported that ANGPT1 oligomers can recruit its receptors to cell–cell contacts, thus forming complexes with angiopoietin-1 receptors from adjoining cells. This process will lead to the activation of phosphatidylinositol 3-kinase and the AKT1 signaling cascades.21 The AKT1 signaling pathway has been implicated as a major contributor to neuronal survival following an ischemic injury to the brain. Akt (protein kinase B) is a subfamily of serine/threonine protein kinase with oncogenic and anti-apoptotic activities.22 Previous research found that the overexpression of neuronal Akt in mice resulted in significant reductions in infarct volumes when compared to wild-type controls.23 Thus, future studies should investigate the effects of ANGPT1 on ischemic stroke via the AKT1 signaling pathway.
One of the advantages of this study is that we used the GEO database to identify potential genes. However, we only selected and characterized one important gene (ANGPT1). GO functional enrichment analysis revealed several other key genes that should be investigated in future, particularly genes that featured in the same functional pathways. For example, apolipoprotein A2 (APOA2) was found to co-exist with ANGPT1 in five functional pathways associated with inflammatory processes (Supplementary Table 1). It is reasonable to assume that APOA2 may also play a role in the occurrence of IS and during subsequent recovery.
There are some limitations to our research that need to be considered. For example, the downregulation of plasma ANGPT1 in IS patients could be attributed to the degradation of mRNA after stroke. However, it is also possible that subjects with lower levels of circulating ANGPT1 are more susceptible to IS. Further studies are now needed to clarify this possibility. In addition, our results were not validated in a separate population with a similar ethnicity. Our results now need to be verified in other ethnic populations; this is because there is a close association between ethnicity and genetic variations. Finally, due to sample size limitations, statistical type 1 errors may have occurred in our study. Thus, future studies could be performed in a more independent manner with a larger sample size.
Conclusions
In this study, we used bioinformatical methods to investigate and characterize a key gene associated with IS (ANGPT1) and its functional SNP (ANGPT1 rs2507799). Our analyses showed that this SNP is associated with an increased risk of IS, a more severe severity of stroke, and a worse outcome. Collectively, our findings might help to improve the future management of ischemic stroke.
Disclosure
None of the authors have any conflicts of interest to declare.
References
1. Lopez AD, Mathers CD. Measuring the global burden of disease and epidemiological transitions: 2002–2030. Ann Trop Med Parasitol. 2006;100(5–6):481–499. doi:10.1179/136485906X97417
2. Boehme AK, Esenwa C, Elkind MS. Stroke risk factors, genetics, and prevention. Circ Res. 2017;120(3):472–495. doi:10.1161/CIRCRESAHA.116.308398
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
4. Wang Y, Zhou L, Chen J, et al. Association of the 3ʹUTR FOXO3a polymorphism rs4946936 with an increased risk of childhood acute lymphoblastic leukemia in a Chinese population. Cell Physiol Biochem. 2014;34(2):325–332. doi:10.1159/000363002
5. Wang W, Li F, Mao Y, et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132(6):641–648. doi:10.1007/s00439-013-1275-6
6. Plasterk RH. Micro RNAs in animal development. Cell. 2006;124(5):877–881. doi:10.1016/j.cell.2006.02.030
7. WHO MONICA Project Principal Investigators. The World Health Organization MONICA Project (monitoring trends and determinants in cardiovascular disease): a major international collaboration. J Clin Epidemiol. 1988;41(2):105–114. doi:10.1016/0895-4356(88)90084-4
8. Zhao J, Wu H, Zheng L, Weng Y, Mo Y. Brain-derived neurotrophic factor G196A polymorphism predicts 90-day outcome of ischemic stroke in Chinese: a novel finding. Brain Res. 2013;1537:312–318. doi:10.1016/j.brainres.2013.08.061
9. Stanne TM, Tjarnlund-Wolf A, Olsson S, Jood K, Blomstrand C, Jern C. Genetic variation at the BDNF locus: evidence for association with long-term outcome after ischemic stroke. PLoS One. 2014;9(12):e114156. doi:10.1371/journal.pone.0114156
10. Quinn TJ, Dawson J, Walters MR, Lees KR. Reliability of the modified Rankin Scale. Stroke. 2007;38(11):
11. Maguire J, Thakkinstian A, Levi C, et al. Impact of COX-2 rs5275 and rs20417 and GPIIIa rs5918 polymorphisms on 90-day ischemic stroke functional outcome: a novel finding. J Stroke Cerebrovasc Dis. 2011;20(2):134–144. doi:10.1016/j.jstrokecerebrovasdis.2009.10.011
12. Golledge J, Clancy P, Maguire J, et al. Plasma angiopoietin-1 is lower after ischemic stroke and associated with major disability but not stroke incidence. Stroke. 2014;45(4):1064–1068. doi:10.1161/STROKEAHA.113.004339
13. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. J Bioinform. 2004;20(3):307–315. doi:10.1093/bioinformatics/btg405
14. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
15. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
16. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res. 1988;16(3):1215. doi:10.1093/nar/16.3.1215
17. Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10(3):165–177. doi:10.1038/nrm2639
18. Chen J, Yang T, Yu H, et al. A functional variant in the 3ʹ-UTR of angiopoietin-1 might reduce stroke risk by interfering with the binding efficiency of microRNA 211. Hum Mol Genet. 2010;19(12):2524–2533. doi:10.1093/hmg/ddq131
19. Zhang ZG, Zhang L, Croll SD, Chopp M. Angiopoietin-1 reduces cerebral blood vessel leakage and ischemic lesion volume after focal cerebral embolic ischemia in mice. Neurisci. 2002;113(3):683–687. doi:10.1016/S0306-4522(02)00175-6
20. Zheng Q, Zhu D, Bai Y, Wu Y, Jia J, Hu Y. Exercise improves recovery after ischemic brain injury by inducing the expression of angiopoietin-1 and Tie-2 in rats. Tohoku J Exp Med. 2011;224(3):221–228. doi:10.1620/tjem.224.221
21. Lee HJ, Cho CH, Hwang SJ, et al. Biological characterization of angiopoietin-3 and angiopoietin-4. FASEB J. 2004;18(11):1200–1208. doi:10.1096/fj.03-1466com
22. Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29(5):233–242. doi:10.1016/j.tibs.2004.03.006
23. Ohba N, Kiryu-Seo S, Maeda M, Muraoka M, Ishii M, Kiyama H. Transgenic mouse overexpressing the Akt reduced the volume of infarct area after middle cerebral artery occlusion. Neurosci Lett. 2004;359(3):159–162. doi:10.1016/j.neulet.2004.02.029
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.