Back to Journals » International Journal of General Medicine » Volume 14
Bioinformatic Exploration for Prognostic Significance of Sphingolipid Metabolism-Related Genes in Invasive Ductal Carcinoma Using the Cancer Genome Atlas Cohort
Authors Kim SJ, Lee JH , Park WJ, Kim S
Received 7 July 2021
Accepted for publication 29 July 2021
Published 12 August 2021 Volume 2021:14 Pages 4423—4434
DOI https://doi.org/10.2147/IJGM.S328376
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Su-Jeong Kim,1,* Jae-Ho Lee,2,* Woo-Jae Park,1 Shin Kim3– 5
1Department of Biochemistry, College of Medicine, Gachon University, Yeonsu-gu, Incheon, 21999, Republic of Korea; 2Department of Anatomy, School of Medicine, Keimyung University, Dalseo-gu, Daegu, 42601, Republic of Korea; 3Department of Immunology, School of Medicine, Keimyung University, Dalseo-gu, Daegu, 42601, Republic of Korea; 4Institute of Medical Science, Keimyung University, Dalseo-gu, Daegu, 42601, Republic of Korea; 5Institute for Cancer Research, Keimyung University Dongsan Medical Center, Dalseo-gu, Daegu, 42601, Republic of Korea
*These authors contributed equally to this work
Correspondence: Shin Kim Email [email protected]
Introduction: Sphingolipid metabolism is a highly controlled process that is involved in regulating bioactive lipid signaling pathways and serves important roles in several cellular processes in breast cancer. Invasive ductal carcinoma (IDC), which is characterized by the malignant proliferation of the ductal epithelium and stromal invasion, is the most common type of breast cancer. Recent advances in genetic research have accelerated the discovery of novel prognostic factors and therapeutic targets for the disease. The aim of the present study was to investigate the expression and prognostic significance of sphingolipid metabolism-related genes in female IDC.
Methods: The present study used gene expression RNAseq data obtained from The Cancer Genome Atlas breast invasive carcinoma (TCGA BRCA) datasets.
Results: Sphingolipid metabolism-related genes exhibited dysregulated mRNA expression levels in IDC. The Student’s t-test revealed that SMPDL3B, B4GALNT1, LPAR2, and LASS2 were significantly upregulated, while LASS3, LPAR1, B4GALT6, GAL3ST1, HPGD, ST8SIA1, UGT8, and S1PR1 were significantly downregulated in female IDC tissues compared with normal solid tissues. Kaplan–Meier survival analyses revealed that high SMPDL3B mRNA expression levels were associated with good prognosis in female IDC, suggesting that SMPDL3B plays a tumor suppressor role. To the best of our knowledge, the present study was the first to report that dysregulated expressions of SMPDL3B are significantly associated with age, estrogen receptor status, progesterone receptor status, and histological subtype.
Conclusion: Taken together, our study indicated that SMPDL3B may have a pathophysiological role and serve as a novel prognostic biomarker in IDC.
Keywords: SMPDL3B, sphingolipid metabolism, invasive ductal carcinoma, TCGA
Introduction
Breast cancer is the most common and life-threatening malignancy in females worldwide.1 Breast carcinoma is the most prevalent malignant type and is classified as carcinoma in-situ and invasive breast cancer.2 Invasive ductal carcinoma (IDC) is the most common type of invasive breast cancer, accounting for up to 80% of diagnosed breast cancer cases.3 There are clinical prognostic biomarkers for breast cancer, including size, histological grade, and estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 status.4 In particular, molecular sub-classification systems such as receptors play an important role in clinical therapeutic strategy.5 However, despite the availability of therapeutic strategies for breast cancer based on molecular subtype, breast cancer is still not overcome.1 Therefore, further studies are required to identify promising molecular biomarkers that can provide new treatment avenues.
Recent advances in genomic profiling using next generation sequencing have made it possible to identify the genetic characteristics of disease, particularly in cancer. Several large-scale cancer genome studies have been conducted, and The Cancer Genome Atlas (TCGA) is a research consortium that may be used to investigate genes in different cancer types.6 Moreover, TCGA may be used to investigate specific histological types of cancer, such as female IDC, by utilizing histological data and clinical parameters.
Sphingolipid metabolism is a highly-regulated intracellular process that controls the synthesis and degradation of bioactive lipids, including ceramide and sphingosine-1-phosphate,7 which plays an important role in biological processes such as angiogenesis, ageing, cancer biology, degenerative diseases, diabetes, immune responses, and inflammation.8 Accumulating evidences revealed that dysregulated sphingolipid metabolism-related genes are implicated in human breast cancer. For example, LAG1 longevity assurance homolog 2 (LASS2) and LASS6 mRNA levels are increased in breast cancer tissues compared with matched normal tissues,9 and overexpressed LASS4 and LASS6 reduced cell proliferation.10 Furthermore, upregulation of sphingosine kinase 1 (SPHK1) was significantly associated with poor prognosis11 and metastasis.12 Moreover, downregulation of 15-hydroxyprostaglandin dehydrogenase (HPGD) has an unfavorable effect on the overall survival of patients with triple negative breast cancer.13 While the altered expression of various sphingolipid metabolism-related genes in breast cancer and their potentials as prognostic factors have been reported in the aforementioned studies, few studies have investigated sphingolipid metabolism-related genes in female IDC.
Therefore, the aim of the present study was to investigate the expression of sphingolipid metabolism-related genes in female IDC, as well as to evaluate their prognostic significance, using gene expression RNAseq data obtained from TGCA breast invasive carcinoma (TCGA BRCA) datasets.
Materials and Methods
Gene Expression Datasets and Cluster Analysis
TCGA BRCA gene expression RNAseq datasets (level 3, dataset ID: TCGA.BRCA.sampleMap/HiSeqV2) and clinical parameters (dataset IDs: TCGA.BRCA.sampleMap/BRCA_clinicalMatrix and survival/BRCA_survival.txt) were downloaded from the UCSC Xena public database (https://xena.ucsc.edu). TCGA BRCA dataset consisted of 1218 samples, including 1097 primary tumor tissues, 7 metastatic tumor tissues, and 114 normal solid tissues (NST). NST were taken from normal tissues adjacent to the tumor. To analyze the RNAseq data of female IDC, female IDC datasets were sorted from TCGA BRCA using clinical parameters. The mRNA expression of the sphingolipid metabolism-related genes14 was identified from the female IDC dataset. In order to identify genes with ≥2-fold changes (2FC) in mRNA expression levels between IDC and NST, the difference between average values of the two groups was calculated, and genes with a value greater than 1 were selected. This study met the publication guidelines for using TCGA datasets (http://www.cancer.gov/about-nci/organization/ccg/research/structrual-genomics/tcga/ using-tcga/citing-tcga). Cluster 3.015 was used for cluster analysis, and samples with statistically similar gene expression were classified into groups. TreeView 1.6 (www.eisenlab.org/eisen) was used to visualize the resulting heat map. The mRNA expression levels of the heat maps were scaled (quantile normalization16 and median-centered) within columns for visualization.
Survival Analysis
Survival data (death event and survival time) were available for 655 female IDC patients. For survival analysis, the mean gene expression value of the selected sphingolipid metabolism-related genes was used as a cutoff to divide the patients into high- and low-expression groups. Survival analysis was performed using the Kaplan–Meier method, and the log rank test was used to identify statistically significant differences between the two groups.
Statistical Analysis
Statistical analysis was performed using SPSS software (version 25.0; IBM SPSS, Armonk, NY, USA). The Kolmogorov–Smirnov test was performed to assess normality. Differences in mRNA expression levels between groups were analyzed using the Student’s t-test. The associations between clinicopathological parameters and the dysregulated sphingolipid metabolism-related genes were analyzed using Chi-square test or Fisher’s exact test for categorical variables. Correlation analysis between inter-individual mRNA expression levels of the sphingolipid metabolism-related genes was performed using the Spearman correlation coefficient analysis for continuous variables. P<0.05 was considered to indicate a statistically significant difference.
Results
Sphingolipid Metabolism-Related Genes are Dysregulated in Female IDC Compared with NST in TCGA BRCA
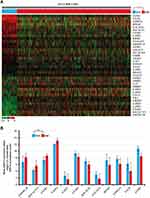
The heat map revealed relative mRNA expression levels of various sphingolipid metabolism-related genes in female IDC tissues and NST (Figure 1). The Student’s t-test revealed that a total of 36 sphingolipid metabolism-related genes were significantly altered in female IDC tissues compared with NST. The results are presented as a heat map (P<0.05; Figure 2A). When a ≥2FC in mRNA expression was used a cutoff, sphingomyelin phosphodiesterase acid like 3B (SMPDL3B), beta-1,4-N-acetyl-galactosaminyltransferase 1 (B4GALNT1), lysophosphatidic acid receptor 2 (LPAR2), and LAG1 longevity assurance homolog 2 (LASS2) were significantly upregulated in female IDC, whereas LAG1 longevity assurance homolog 3 (LASS3), lysophosphatidic acid receptor 1 (LPAR1), beta-1,4-galactosyltransferase 6 (B4GALT6), galactose-3-O-sulfotransferase 1 (GAL3ST1), HPGD, ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 (ST8SIA1), UDP glycosyltransferase 8 (UGT8), sphingosine-1-phosphate receptor 1 (S1PR1) were significantly downregulated (P<0.001, |2FC| ≥1.0; Figure 2B), compared with NST. These results suggested that the aforementioned 12 dysregulated sphingolipid metabolism-related genes may play a crucial role in pathophysiology of female IDC.
KM Survival Analysis Identified SMPDL3B as a Prognostic Biomarker in Female IDC
Among the altered sphingolipid metabolism-related genes in female IDC, the KM survival analysis and Log rank test demonstrated that higher SMPDL3B mRNA expression levels were found to be associated with favorable overall survival in female IDC (Figure 3, P=0.003).
Altered mRNA Expression Levels of Sphingolipid Metabolism-Related Genes are Associated with Clinicopathological Parameters in Female IDC
To investigate the clinicopathological implications of the dysregulated sphingolipid metabolism-related genes, Chi-square test or Fisher’s exact test were performed. The clinicopathological characteristics of female IDC patients of TCGA BRCA are presented in Tables 1 and 2. The SMPDL3B mRNA expression level was significantly associated with age, estrogen receptor (ER) status, progesterone receptor (PR) status, and histological subtypes according to immunohistochemistry (IHC) (P=0.002, P<0.001, P<0.001, and P<0.001, respectively, Table 1). The B4GALNT1 mRNA expression level was significantly associated with T stage, ER status, PR status, and histological subtype according to IHC (P=0.004, P<0.001, P<0.001, and P<0.001, respectively, Table 1). The LPAR2 mRNA expression level was significantly associated with T stage, ER status, PR status, and histological subtype according to IHC (P<0.001, P<0.001, P<0.001, and P<0.001, respectively, Table 1). The LASS2 mRNA expression level was significantly associated with age, ER status, PR status, and histological subtype according to IHC (P=0.003, P<0.001, P<0.001, and P<0.001, respectively, Table 1). Moreover, the GAL3ST1 mRNA expression level was significantly associated with ER status, PR status, and histological subtype according to IHC (P=0.003, P=0.011, and P<0.001, respectively, Table 2). The LASS3 mRNA expression level was significantly associated with age and histological subtype according to IHC (P=0.036 and P=0.041, respectively, Table 2). The B4GALT6 mRNA expression level was significantly associated with ER status, PR status, epidermal growth factor receptor type 2 (HER2), and histological subtype according to IHC (P=0.006, P<0.001, P=0.003, and P<0.001, respectively, Table 2). The HPGD mRNA expression level was significantly associated with age, ER status, PR status, and histological subtype according to IHC (P=0.024, P<0.001, P=0.007, and P<0.001, respectively, Table 2). The UGT8 mRNA expression level was significantly associated with age, N stage, ER status, PR status, HER2 and histological subtype according to IHC (P=0.003, P=0.029, P<0.001, P<0.001, P=0.003, and P<0.001, respectively, Table 2). The ST8SIA1 mRNA expression level was significantly associated with age, T stage, ER status, PR status, and histological subtype according to IHC (P=0.019, P=0.008, P<0.001, P<0.001, and P<0.001, respectively, Table 2). The S1PR1 mRNA expression level was significantly associated with M stage and histological subtype according to IHC (P=0.005 and P<0.001, respectively, Table 2). The LPAR1 mRNA expression level was significantly associated with T stage, ER status, PR status, and histological subtype according to IHC (P<0.001, P<0.001, P<0.001, and P<0.001, respectively, Table 1). Interestingly, we found that lower expression levels of SMPDL3B were 2.676 times more frequent in ER-positive IDC than in ER-negative (odds ratio (OR), 2.676; 95% confidence interval (CI), 1.821 to 3.932; P<0.001). Moreover, we found that lower expression levels of SMPDL3B were 1.898 times more frequent in PR-positive IDC than in PR-negative (OR, 1.898; 95% CI, 1.357 to 2.655; P<0.001).
 |
Table 1 Upregulated mRNA Expression Levels of Sphingolipid Metabolism-Related Genes in Relation to Clinicopathological Parameters of IDC |
 |
Table 2 Downregulated mRNA Expression Levels of Sphingolipid Metabolism-Related Genes in Relation to Clinicopathological Parameters of IDC |
Altered mRNA Expression Levels of Sphingolipid Metabolism-Related Genes Have Inter-Individual Correlations in Female IDC
Spearman correlation coefficient analysis was performed to explore the correlation among the significantly altered sphingolipid metabolisms-related genes in female IDC tissues. Prior to the correlation analysis, the mRNA expression levels of the 12 significantly dysregulated sphingolipid metabolism-related genes in female IDC samples were retrieved from TCGA BRCA cohort. The resulting analysis uncovered 43 significant correlations among the mRNA expression levels of specific sphingolipid metabolism-related genes in female IDC samples (Table 3).
 |
Table 3 Spearman Correlation Analysis Between Inter-Individual Components of Sphingolipid Metabolism-Related Genes |
Discussion
Sphingolipid metabolism regulates diverse biological processes, including breast cancer pathophysiology.17 IDC of the breast cancer, arises from epithelial cells in the inner lining of the milk ducts, and is the most common type of invasive breast cancer that microscopically penetrates through the epithelium of the ducts into the stroma in breast tissue.2,3 As the importance of precision medicine becomes apparent, the number of studies using genomic profiles to identify novel prognostic and therapeutic targets is steadily increasing.18,19 Although methods for precisely targeting or imaging such as peptide-based SPECT radiopharmaceuticals have been studied using the discovered target for diagnosis or treatment of IDC,20–22 its prognosis is still unfavorable. In the present study, gene expression RNAseq data from TCGA BRCA dataset were used to evaluate whether sphingolipid metabolism-related genes are significantly dysregulated, and whether they may serve as potential prognostic indicators in female IDC.
The present study investigated altered genes with ≥2FC in mRNA expression levels in IDC compared with NST. SMPDL3B, B4GALNT1, LPAR2, and LASS2 were significantly upregulated and the other genes (LASS3, LPAR1, B4GALT6, GAL3ST1, HPGD, ST8SIA1, UGT8, and S1PR1) were significantly downregulated (Figure 2). Among the dysregulated sphingolipid metabolism-related genes, only SMPDL3B had prognostic significance in female IDC (Figure 3). SMPDL3B is one of the lipid raft enzymes that regulates lipid composition and fluidity in the plasma membrane of macrophages.23 Interestingly, SMPDL3B modulates podocyte migration and apoptosis, and activates integrin in podocytes.24 Moreover SMPDL3B inversely regulates ceramide-1-phosphate (C1P) levels by interacting with ceramide kinase (CERK) in human podocytes.25 In addition, it has been reported that C1P is involved in enhancement of cancer cell growth, migration, and survival.26 However, CERK was downregulated in IDC (Figure 1), suggesting SMPDL3B and C1P are regulated differently in IDC and podocytes. Notably, a recent report demonstrated that increased SMPDL3B was associated improved prognosis in prostate cancer.27 The present study demonstrated that increased mRNA expression levels of SMPDL3B is significantly associated with improved prognosis (Figure 3). These results suggested that SMPDL3B may function as a tumor suppressor gene.
In the present study, we found that SMPDL3B and LPAR2 were upregulated (Figure 2B), and a significant positive correlation was identified between the genes (Table 3). Additionally, higher LPAR2 mRNA expression levels tended to show favorable overall survival in female IDC (Figure 3). However, LPAR2 showed aggressive characteristics of gastric cancer cells, such as inducing cell migration28 and LPAR2-KO had significantly suppressed the invasion of ovarian cancer cells.29 Although more studies on the mechanism of each gene in IDC are needed, the aforementioned results suggested that SMPDL3B and LPAR2 have different regulatory mechanisms and functions. On the other hand, the downregulated sphingolipid metabolism-related genes such as HPGD, S1PR1, and LPAR1, which are inversely correlated with the SMPDL3B, have been identified (Table 3). Additionally, higher HPGD mRNA expression levels tended to indicate a favorable prognosis (Figure 3). However, it was previously shown that HPGD was significantly upregulated and higher HPGD is significantly associated with a poor overall survival and the increased risk of disease relapse in breast cancer.30 In order to determine the dysregulation and function of HPGD in breast cancer, it is necessary to analyze it by breast cancer subtype.
Recent studies reported significant correlations among sphingolipid metabolism-related genes, such as ceramide synthases. For example, significant correlations were determined between LASS2 and LASS4/LASS6, and between LASS4 and LASS6 in breast cancer.9 Moreover, significant correlations between LASS2 and LASS4, and between LASS5 and LASS4/LASS6 were identified in various colorectal cancer cohorts.31 Interestingly, in the present study, only the correlation between LASS2 and LASS3 was significant in female IDC (Table 3).
Although further studies are required to elucidate the underlying mechanisms involved in the inter-individual correlation analysis in female IDC, our results suggested that networks of sphingolipid metabolism-related genes may be implicated in the pathophysiology of the disease.
Conclusions
To the best of our knowledge, the present study was first to identify the significantly dysregulated sphingolipid metabolism-related gene, SMPDL3B as a novel prognostic biomarker for female IDC using TCGA BRCA datasets. Our results suggested that SMPDL3B plays an important role in the pathophysiology of female IDC, and may serve as a potential prognostic biomarker as well as promising therapeutic target for the disease.
Funding
This work was supported by the research promoting grant from the Keimyung University Dongsan Medical Center in 2014.
Disclosure
The authors report no conflicts of interest in this work.
References
1. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi:10.3322/caac.21412
2. Makki J. Diversity of Breast Carcinoma: histological Subtypes and Clinical Relevance. Clin Med Insights Pathol. 2015;8:23–31. doi:10.4137/CPath.S31563
3. Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010;1(2):109–126.
4. Bundred NJ. Prognostic and predictive factors in breast cancer. Cancer Treat Rev. 2001;27(3):137–142. doi:10.1053/ctrv.2000.0207
5. Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–35. doi:10.1016/j.breast.2015.07.008
6. Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol. 2016;1418:111–141. doi:10.1007/978-1-4939-3578-9_6
7. Pralhada Rao R, Vaidyanathan N, Rengasamy M, Mammen Oommen A, Somaiya N, Jagannath MR. Sphingolipid metabolic pathway: an overview of major roles played in human diseases. J Lipids. 2013;2013:178910. doi:10.1155/2013/178910
8. Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. doi:10.1038/nrm2329
9. Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem Biophys Res Commun. 2010;391(1):219–223. doi:10.1016/j.bbrc.2009.11.035
10. Hartmann D, Lucks J, Fuchs S, et al. Long chain ceramides and very long chain ceramides have opposite effects on human breast and colon cancer cell growth. Int J Biochem Cell Biol. 2012;44(4):620–628. doi:10.1016/j.biocel.2011.12.019
11. Geffken K, Spiegel S. Sphingosine kinase 1 in breast cancer. Adv Biol Regul. 2018;67:59–65. doi:10.1016/j.jbior.2017.10.005
12. Acharya S, Yao J, Li P, et al. Sphingosine Kinase 1 Signaling Promotes Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2019;79(16):4211–4226. doi:10.1158/0008-5472.CAN-18-3803
13. Kochel TJ, Goloubeva OG, Fulton AM. Upregulation of Cyclooxygenase-2/Prostaglandin E2 (COX-2/PGE2) Pathway Member Multiple Drug Resistance-Associated Protein 4 (MRP4) and Downregulation of Prostaglandin Transporter (PGT) and 15-Prostaglandin Dehydrogenase (15-PGDH) in Triple-Negative Breast Cancer. Breast Cancer (Auckl). 2016;10:61–70. doi:10.4137/BCBCR.S38529
14. Ruckhaberle E, Rody A, Engels K, et al. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res Treat. 2008;112(1):41–52. doi:10.1007/s10549-007-9836-9
15. de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi:10.1093/bioinformatics/bth078
16. Evans C, Hardin J, Stoebel DM. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions. Brief Bioinform. 2018;19(5):776–792. doi:10.1093/bib/bbx008
17. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi:10.1038/nrc.2017.96
18. Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747–756. doi:10.1038/nrc4015
19. Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer–limitations and solutions. Nat Rev Clin Oncol. 2015;12(12):693–704. doi:10.1038/nrclinonc.2015.123
20. Ahmadpour S, Noaparast Z, Abedi SM, Hosseinimehr SJ. (99m)Tc-(tricine)-HYNIC-Lys-FROP Peptide for Breast Tumor Targeting. Anticancer Agents Med Chem. 2018;18(9):1295–1302. doi:10.2174/1871520618666180307142027
21. Ahmadpour S, Noaparast Z, Abedi SM, Hosseinimehr SJ. (99m)Tc-HYNIC-(tricine/EDDA)-FROP peptide for MCF-7 breast tumor targeting and imaging. J Biomed Sci. 2018;25(1):17. doi:10.1186/s12929-018-0420-x
22. Ahmadpour S, Hosseinimehr SJ. Recent developments in peptide-based SPECT radiopharmaceuticals for breast tumor targeting. Life Sci. 2019;239:116870. doi:10.1016/j.lfs.2019.116870
23. Heinz LX, Baumann CL, Koberlin MS, et al. The Lipid-Modifying Enzyme SMPDL3B Negatively Regulates Innate Immunity. Cell Rep. 2015;11(12):1919–1928. doi:10.1016/j.celrep.2015.05.006
24. Yoo TH, Pedigo CE, Guzman J, et al. Sphingomyelinase-like phosphodiesterase 3b expression levels determine podocyte injury phenotypes in glomerular disease. J Am Soc Nephrol. 2015;26(1):133–147. doi:10.1681/ASN.2013111213
25. Mallela SK, Mitrofanova A, Merscher S, Fornoni A. Regulation of the amount of ceramide-1-phosphate synthesized in differentiated human podocytes. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(12):158517. doi:10.1016/j.bbalip.2019.158517
26. Hait NC, Maiti A. The Role of Sphingosine-1-Phosphate and Ceramide-1-Phosphate in Inflammation and Cancer. Mediators Inflamm. 2017;2017:4806541. doi:10.1155/2017/4806541
27. Waldbillig F, Nitschke K, Abdelhadi A, et al. Phosphodiesterase SMPDL3B Gene Expression as Independent Outcome Prediction Marker in Localized Prostate Cancer. Int J Mol Sci. 2020;21(12):4373. doi:10.3390/ijms21124373
28. Yang D, Yang W, Zhang Q, Hu Y, Bao L, Damirin A. Migration of gastric cancer cells in response to lysophosphatidic acid is mediated by LPA receptor 2. Oncol Lett. 2013;5(3):1048–1052. doi:10.3892/ol.2013.1107
29. Onallah H, Davidson B, Reich R. Diverse Effects of Lysophosphatidic Acid Receptors on Ovarian Cancer Signaling Pathways. J Oncol. 2019;2019:7547469. doi:10.1155/2019/7547469
30. Lehtinen L, Vainio P, Wikman H, et al. 15-Hydroxyprostaglandin dehydrogenase associates with poor prognosis in breast cancer, induces epithelial-mesenchymal transition, and promotes cell migration in cultured breast cancer cells. J Pathol. 2012;226(4):674–686. doi:10.1002/path.3956
31. Jang SW, Park WJ, Min H, et al. Altered mRNA expression levels of the major components of sphingolipid metabolism, ceramide synthases and their clinical implication in colorectal cancer. Oncol Rep. 2018;40(6):3489–3500. doi:10.3892/or.2018.6712
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.