Back to Journals » International Journal of Nanomedicine » Volume 12
Biodegradable and biocompatible cationic polymer delivering microRNA-221/222 promotes nerve regeneration after sciatic nerve crush
Authors Song J , Li X, Li Y, Che J, Li X, Zhao X, Chen Y , Zheng X, Yuan W
Received 11 January 2017
Accepted for publication 3 April 2017
Published 2 June 2017 Volume 2017:12 Pages 4195—4208
DOI https://doi.org/10.2147/IJN.S132190
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Jialin Song,1,2 Xueyang Li,3 Yingli Li,4,5 Junyi Che,6 Xiaoming Li,6 Xiaotian Zhao,6 Yinghui Chen,7,* Xianyou Zheng,1,* Weien Yuan6,*
1Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, 2Department of Orthopedics, Shanghai University of Medicine and Health, Shanghai, Sixth People’s Hospital East Campus, Shanghai, 3Department of Plastic and Reconstructive Surgery, Xuzhou Medical College Affiliated Hospital, Xuzhou, Jiangsu, 4Department of Plastic Surgery, The General Hospital of Jinan Military Command, Jinan, Shandong, 5Department of Plastic Surgery, Chang Hai Hospital, Second Military Medical University, 6School of Pharmacy, Shanghai Jiao Tong University, 7Department of Neurology, Jinshan Hospital, Fudan University, JinShan District, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: MicroRNA (miRNA) has great potential to treat a wide range of illnesses by regulating the expression of eukaryotic genes. Biomaterials with high transfection efficiency and low toxicity are needed to deliver miRNA to target cells. In this study, a biodegradable and biocompatible cationic polymer (PDAPEI) was synthetized from low molecular weight polyethyleneimine (PEI1.8kDa) cross-linked with 2,6-pyridinedicarboxaldehyde. PDAPEI showed a lower cytotoxicity and higher transfection efficiency than PEI25kDa in transfecting miR-221/222 into rat Schwann cells (SCs). The upregulation of miR-221/222 in SCs promoted the expression of nerve growth factor and myelin basic protein in vitro. The mouse sciatic nerve crush injury model was used to evaluate the effectiveness of PDAPEI/miR-221/222 complexes for nerve regeneration in vivo. The results of electrophysiological tests, functional assessments, and histological and immunohistochemistry analyses demonstrated that PDAPEI/miR-221/222 complexes significantly promoted nerve regeneration after sciatic nerve crush, specifically enhancing remyelination. All these results show that the use of PDAPEI to deliver miR-221/222 may provide a safe therapeutic means of treating nerve crush injury and may help to overcome the barrier of biomaterial toxicity and low efficiency often encountered during medical intervention.
Keywords: miR-221/222, PDAPEI, nerve regeneration, remyelination
Introduction
Peripheral nerve crush injury is relatively common in clinical situations, arising from trauma and often leads to severe motor and sensory dysfunctions.1,2 A number of therapies have been investigated for nerve regeneration including the use of growth factors, tissue engineering scaffolds, and physical stimulation; all have been demonstrated to result in relatively satisfying repair outcome.3,4 Gene therapy offers a new advancement that may contribute in the treatment of nerve crush injury via the delivery of specific genetic materials (DNA and RNA) to the targeted cells. The gene delivery system, containing viral vectors, nonviral vectors, and engineered vectors, provides the means for transporting genetic material into the target cells.5–14 Nonviral vectors have been thoroughly investigated in the past few years due to their lower immunogenicity and cytotoxicity, higher transfection efficiency and longer gene expression duration.6,15,16 Polyethylenimine (PEI) is considered the gold standard for both in vivo and in vitro gene transfer in the field of cationic polymers because of its strong DNA condensation capacity and specific endosomolytic activity.17–20 Low molecular weight PEI (<20 kDa) has shown relatively low cytotoxicity but poor transfection efficiency.21,22 Degradable linkers can conjugate with low molecular weight PEI to form cationic polymers with lower cytotoxicity and higher transfection efficiency. Lots of works have been done about cross-linked low molecular weight PEI in our previous research.23,24 In this study, we cross-linked PEI1.8kDa with 2,6-pyridinedicarboxaldehyde (PDA) to develop a biodegradable low molecular weight cationic polymer (PDAPEI), which showed a higher transfection efficiency and a relatively low cytotoxicity in various cell lines.
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression of target mRNAs to inhibit or induce cell differentiation, proliferation, migration, apoptosis, and even metabolism.25–27 Many miRNAs have been shown to be relevant in nerve regeneration. MiR-338 and miR-21 cotransfection can accelerate axonal regeneration and facilitate functional recovery following peripheral nerve injury.28 MiR-1 regulates Schwann cell (SC) proliferation and migration by modulating brain-derived neurotrophic factor following peripheral nerve injury.29 MiR-431 regulates axon regeneration in mature sensory neurons via the Wnt antagonist Kremen1.30 MiR-221/222 plays an important role in promoting SC proliferation, migration, and regulating nerve growth factor (NGF) expression through several signaling pathways.31,32 However, its role in peripheral nerve regeneration is still not clear. SCs play a crucial role in peripheral nerve regeneration by promoting myelination and neurotrophic function.33 Because the sciatic nerve crush injury (SNCI) model is widely used for studying peripheral nerve regeneration, we selected SCs for in vitro study and SNCI for the in vivo study.
In previous work, we have synthesized an efficient and nontoxic biological response carrier named PDAPEI.34,35 In this study, we examined miR-221/222 upregulating the gene for peripheral nerve regeneration. PDAPEI was used as the vector and its characteristics, cytotoxicity, and transfection efficiency were investigated in SCs. To further understand the potential for nerve regeneration, we explored the expression of NGF and myelin basic protein (MBP) in vitro in SCs following transfection and the regeneration of SNCI followed PDAPEI/miR-221/222 complexes treatment in vivo.
Materials and methods
Materials
PEI (25 and 1.8 kDa) and anhydrous ethylene dichloride (EDC) were purchased from Sigma-Aldrich (St Louis, MO, USA). PDA was purchased from TCI (Shanghai) Development Co., Ltd. Cell Counting Kit (CCK-8) was purchased from Sigma-Aldrich (Milwaukee, WI, USA). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (MediaTech, Herndon, VA, USA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA). Mir-221/222 encoding GFP was constructed by Bioroot Biology (Shanghai, China) with the following sequences: 3′-UUUGGUCGUCUGUUACAUCG-5′ and 3′-UGGGUCAUCGGUCUACAUCG-3′.
Cell culture
Rat RSC96 SCs were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM containing 10% FBS and 1% antibiotics at 37°C with a CO2 concentration of 5%. Immunofluorescence staining of SCs makers of p75NGFR and S100-β (Abcam, Cambridge, UK) demonstrated that highly purified SCs (>99%) were used in our experiment (Figure 1).
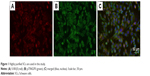  | Figure 1 Highly purified SCs are used in this study. |
Preparation of PDAPEI
PDAPEI cationic polymer synthesis was performed as previously described in our published paper.34 Briefly, PEI1.8kDa (1 mmol) was introduced into anhydrous EDC solution (20 mL) and stirred vigorously to dissolve completely. PDA (2 mmol) was dissolved in anhydrous EDC (20 mL) and then added dropwise into the PEI solution with vigorous stirring at room temperature. After stirring for 48 h, the solvent was evaporated. After 24 h of dialysis to remove small fragments (<10 kDa), the freeze-dried yellowish dialysis was PDAPEI. The polymer was stored at −20°C for further use. The structure of PDAPEI was confirmed by proton nuclear magnetic resonance (1H NMR) and Fourier transform infrared spectroscopy (FTIR).
Preparation and characterization of PDAPEI complexes (polyplexes)
PDAPEI/miR-221/222 complexes were prepared by adding different concentrations of PDAPEI (2 mg/mL) to the DNA solution (20 ng/μL, miR-221 and miR-222 of equal weight). In this study, we selected preset nitrogen-to-phosphor (N/P) trial ratios of PDAPEI/miR-221/222 and used PEI25kDa/miR-221/222 complexes as a control. The particle size and zeta potential of the polyplexes were measured, using Brookhaven Particle Size Analyzer (90 Plus). The morphology of the polyplexes was observed using a transmission electron microscope (TEM, JEM 2010 system JEOL, Japan). The condensation ability of PDAPEI/miR-221/222 complexes was evaluated by gel electrophoresis, stained with ethidium bromide for semiquantitative measurements, and detected using a Gel Doc EZ system (Bio-Rad, Hercules, CA, USA). The results were repeated three times and the densitometry was shown as mean ± standard deviation (SD).
Cytotoxicity of the PDAPEI/miR-221/222 nanoparticles
In vitro cytotoxicity was determined using CCK-8 assay. SCs (1×104/well) were seeded into a 96-well plate and incubated at 37°C for 24 h. Then the solutions of PDAPEI/miR221/222 with different N/P ratios were added into the wells (PEI25kDa/miR-221/222 complexes as control). After 4 and 24 h of incubation, 10 μL CCK8 reagent was added into each well, and the plates were further incubated for 2 h for cell viability determination. A multifunctional microplate reader (SpectraMax M3 Multi-Mode Microplate Reader) was used to measure cell viability. In vivo toxicity was determined by visceral and local muscle tissue damage following local right thigh muscle injection of the PDAPEI/miR221/222 complexes (at N/P ratio of 300, saline as control). The main organs and right thigh muscle were harvested 1 week after injection to determine the damage by hematoxylin–eosin staining.
Gene transfection efficiency
To determine the transfection efficiency of PDAPEI/miR221/222 complexes, SCs (5–10×104/mL) were seeded into 12-well plates and cultured for 24 h till the cells reached 80%–90% confluence. The medium in each well was then replaced with 200 μL of PDAPEI/miR221/222 complexes solution at different N/P ratios and 800 μL DMEM for an additional 4 h of incubation (naked miR-221/222 as negative control, PEI25kDa/miR-221/222 at N/P ratio of 10 as positive control). The transfection medium was replaced with 1 mL of fresh DMEM medium containing 10% FBS and incubated for another 48 h. Then GFP-positive cells were viewed under a fluorescence microscope (Olympus, Tokyo, Japan) and quantified by flow cytometry (BectoneDickinson, Franklin Lakes, NJ, USA).
NGF and MBP expression
SCs in 12-well plates were divided into four groups: miR-221/222+ group (PDAPEI transfected miR-221/222 mimics), miR-221/222− group (PDAPEI transfected miR-221/222 inhibitor), negative control group (PDAPEI without miRNA), and the blank group not transfected with anything (PBS control). Western blot analysis was performed to examine NGF and MBP expression. SCs were cultured for an additional 4 days after transfection. The treated SCs were washed in 0.1 M PBS on ice and lysed in RIPA buffer including 0.005 M Tris, 0.001 M ethylenediaminetetraacetic acid, 100 μg/mL PMSF, and 1 mM activated sodium orthovanadate. The solution was then centrifuged at 1,500 rpm for 15 min, and the supernatants were collected. Protein concentrations were determined using a bicinchoninic acid assay kit (Beyotime), according to the manufacturer’s protocol. The antibodies used in this procedure were as follows: anti-rabbit MBP (1:1,000; Abcam, Cambridge, MA, USA), anti-rabbit NGF (1:1,000; Abcam) as primary antibodies and horseradish peroxidase (HRP)-labeled secondary antibody (1:3,000; Santa Cruz Biotech, Santa Cruz, CA, USA). Western blotting was performed at least twice for each sample and quantified using ImageJ software. For gene expression studies, cells were collected after 4 days of incubation following transfection. Total RNA was isolated from the cells using a High Pure RNA isolation kit (Qiagen, Valencia, CA, USA). The RNA was reverse transcribed into double-stranded cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The primer sequences of NGF (forward: 5′-CATGCTGGACCCAAGCTCA-3′; reverse: 5′-GACATTACGCTATGCACCTCAGTG-3′) and MBP (forward: 5′-GGCCCCGTGGATGGA-3′; reverse: 5′-GAGGCGCGAAAGGAGATG-3′) were synthesized and purified by TAKARA, Inc. The cDNAs for NGF and MBP were amplified by RT-PCR under the following conditions: denaturation at 95°C for 10 min; 40 cycles of denaturation steps at 95°C for 15 s per cycle, and annealing/extending processes at 60°C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control and the agarose gels electrophoresis results were visualized under UV light.
Animal study
Twenty-four Sprague-Dawley rats (male, weighing 200–250 g) were housed under standard laboratory conditions. The rats were anesthetized by intraperitoneal injection of 50 mg/kg pentobarbital sodium, and a 2-cm incision was made on the right thigh to expose the sciatic nerve under sterile conditions. The sciatic nerve was crushed with a vascular clamp for 30 seconds to cause constant SNCI. The incision was then closed with a suture. The animals were randomly divided into four groups (n=6): the PDAPEI/miR-221/222+ complexes group, the PDAPEI/miR-221/222− complexes group, the negative control group (PDAPEI without miRNA), and the blank group (treated with saline without PDAPEI or miRNA). The treatment was commenced 7 days after the injury when SNCI model was successfully made and the wound had healed completely. Different solutions were injected into around the injury site once daily for 7 days in all four groups. The PDAPEI/miR221/222 (mimics or inhibitor) (N/P ration of 200) group was injected with 100 μL of solution and the total amount of miR-221/222 injected was 10 μg for each mouse. The PDAPEI without miRNA group was injected with 300 μg PDAPEI in 100 μL of saline, whereas the control group was injected with 100 μL of saline alone. All rats were housed under standardized laboratory conditions and monitored to observe any changes in ordinary conditions and activities. All animal experiments were conducted strictly in accordance with the guidelines approved by the Regulations for the Administration of Affair Concerning Laboratory Animals for Shanghai Jiao Tong University, the National Institutes of Health Guide for Care and Use of Laboratory Animals (GB14925-2010), the Regulations for the Administration of Affairs Concerning Experimental Animals (China, 2014). The Animal Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital reviewed and approved the experiments.
Electrophysiological, sciatic function index (SFI), and the weight of triceps surae muscles were used to evaluate the functional recovery in the four groups. Each rat was anesthetized and the sciatic nerve was exposed. Then a monopolar recording electrode and bipolar stimulating electrodes were placed to induce and record the electrical activity. A digital MYTO electromyograph machine (Esaote, Genoa, Italy) was used to evaluate the nerve conduction velocity (NCV) and distal compound motor action potential (DCMAP). The SFI was calculated to assess the recovery of nerve function by the walking track analysis at the same time. Meanwhile, triceps surae muscles were carefully resected and weighted to evaluate the recovery rate of triceps surae muscles. All procedures used were described in our previous study.36
To investigate the extent of nerve remyelination, we assessed the ultrastructure of myelin sheaths under a TEM and MBP expression by immunohistochemistry staining. A 2-mm segment was harvested from the injury site and fixed in cold buffered 3% glutaraldehyde solution then processed for TEM (JEOL JEM 2010 system). The captured images were analyzed with OpenLab software (Improvision, Conventry, UK). For MBP immunohistochemistry analysis, 2-mm thick segments were fixed in 10% neutral buffered formalin for 48 h. After washing, dehydration, and xylene transparent, tissues were embedded in paraffin and cut into 4-μm sections and baked at 65°C for 30 min. The sections were deparaffinized and rehydrated before antigen retrieval using 0.01 M sodium citrate buffer solution under high pressure for 15 min. Sections were washed three times with 0.02 M PBS (3 min for each time) after cooling naturally for antigen repair. The sections were treated with 3% H2O2 and primary anti-MBP antibody (1:100; Sigma-Aldrich) and then incubated at 4°C overnight. HRP-labeled secondary antibody (1:200; Abcam, UK) was added, and the sections were incubated for another 30 min at room temperature. DAB stain and hematoxylin stain were used after incubation. A light microscope was used to visualize and quantify the positive areas. HPIAS-1000, a high-resolution pathological image analysis system was used for the quantitative analysis of MBP expression.37 Investigators were blinded for image analyses and five randomly observation regions were selected in each section.
Statistical analysis
Data are presented as the mean ± standard error of five replicates. Analysis of variance and Student’s t-test were used for statistical analysis, and P<0.05 was considered statistically significant.
Results
Preparation of PDAPEI
The chemical synthesis reaction to produce PDAPEI is shown in Figure 2. The structure of PDAPEI was confirmed by 1H NMR and FTIR, as described in our previous study.34,35
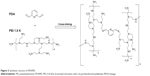  | Figure 2 Synthesis reaction of PDAPEI. |
Characteristics of PDAPEI/miR-221/222 complexes (polyplexes)
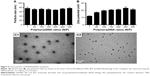
To confirm the characteristics of the polyplexes, agarose gel electrophoresis was performed, and the particle size, zeta potential, and morphology of PDAPEI/miR-221/222 polyplexes were measured. As shown in Figure 3A, the electrophoresis band for miR-221/222 became completely invisible when the N/P reached 5 for PDAPEI, while a partly migration of miR-221/222 was observed at N/P ratio of 4 and below. The results from Gel Doc EZ system detection indicated that the binding capability of PDAPEI gradually increased with a higher N/P ratio and formed an approximate sigmoidal curve (Figure 3B). For the particle size, PDAPEI/miR-221/222 polyplexes at N/P ratios ranging from 1 to 300 were measured and the results are presented in Figure 4A. The particle size of PDAPEI/miR-221/222 was ~70 nm for all 7 ratios, which is almost the same as the size of the PEI25kDa/miR-221/222 particle. When the N/P ratio increased from 1 to 300, the zeta potential of PDAPEI/miR-221/222 had a stable constant positive charge between 27 and 30 mV (Figure 4B). The morphology of PDAPEI/miR-221/222 was measured by TEM, where 60–80 nm diameter particles with spherical shape and uniform size were observed, which was consistent with the particle size analysis (Figure 4C1, C2). The above studies were repeated at 1 month post-PDAPEI synthesis and stored at −20°C and got similar results, which indicated that the polymer is stable under this condition at least 1 month.
Cytotoxicity to SCs and toxicity in vivo
To measure the cytotoxicity of complexes against SCs in vitro, cell counting was performed using CCK-8. As shown in Figure 5, the cytotoxicity of PDAPEI/miR-221/222 to SCs was significantly lower than that of the PEI25kDa/miR 221/222 complexes at 4 and 24 h posttransfection. The PDAPEI/miR-221/222 complexes maintained low cytotoxicity even at increasing concentrations. The viability of cells treated with PDAPEI/miR-221/222 complexes at different N/P ratios was higher than 80%. However, the cytotoxicity of the PEI25kDa/miR-221/222 complexes increased at higher polymer concentrations, at which the cell viabilities were only 25% at a N/P ratio of 200. No obvious cytotoxicity was observed for the naked miR-221/222 group. Interestingly, the pure polymer showed a little higher (no significant difference) cytotoxicity than the polyplexes at the same concentration (both PEI25kDa and PDAPEI), which indicated that the DNA-binding process may decreased the cytotoxicity of the polymer. To test the toxicity in vivo, PDAPEI/miR-221/222 at a N/P ratio of 300 was injected locally into right thigh muscle and showed a negligible toxicity comparable to the saline treated control group. Thus, this new cationic polymer PDAPEI is seemingly biocompatible as desired (Figure 6).
Transfection efficiency to SCs
The miR-221/222 encoding a GFP reporter gene can express GFP 48 h after successful transfer into cells. In our study, different ratios of PDAPEI/miR-221/222 complexes at ratios ranging from 1 to 300 (N/P) were used to transfect SCs. The qualitative result was observed under fluorescence, while quantitative evaluation was performed by flow cytometry. As shown in Figure 7A–D, hardly any fluorescence was observed in miR-221/222 alone group and the PDAPEI/miR-221/222 complexes group showed a considerably high GFP protein expression. Transfection efficiency of the PDAPEI/miR-221/222 complexes became increased as the N/P ratio rose from 1 to 200 and was maintained in the higher ratios from 200 to 300, where a ~15% GFP-positive cells were observed. We also examined the transfection efficiency of PEI25kDa at lower concentrations where cell viability is more than 50% according Figure 5. These results demonstrated that a similar (even better) transfection efficiency of PDAPEI was observed, especially at the N/P ratios of 100 and above, when compared with the positive control PEI25kDa/miR-221/222 complexes (Figure 7E).
NGF and MBP expression
Western blot analysis was used to detect the intracellular expression of NGF and MBP. As shown in Figure 8A, there was no obvious difference between the blank and negative control (NC ) group, indicating that PDAPEI alone did not significantly alter the expression of NGF and MBP in SCs (P>0.05). The relative expression of NGF of miR-221/222+ group was significantly higher than that of the miR-221/222− group (P<0.01). For the expression of MBP, the difference was more obvious between the miR-221/222+ and miR-221/222− group (P<0.001). The effect of miR-221/222 on the expression of NGF and MBP miRNA in SCs was examined by real-time RT-PCR analysis, and the results were in agreement with the results of the Western blot analysis (Figure 8B).
Animal study
In the in vivo study, no severe complications such as inflammation, infection, and anesthesia accident were observed throughout the experiment.
The results of functional recovery are shown in Figure 9. The NCV and DCMAP in the miR-221/222+ group (69.2±3.6 ms−1, 17.4±1.6 mV) were significantly better than those in the other three groups (n=6, P<0.01). At 4 weeks posttreatment, the SFI of the blank group (−42.4±2.7) was significantly lower than that for the miR-221/222+ group (−12.1±2.2) (n=6, P<0.05). The recovery rates of triceps surae muscles in the miR-221/222+ group were significantly higher than those in the NC group and the blank group (n=6, P<0.05). The functional recovery results for the miR-221/222− group were the worst compared to the other three groups.
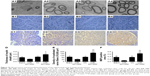
The ultrastructure and immunohistochemistry of the regenerated nerve 4 weeks post-treatment are shown in Figure 10. The myelin sheath thickness was measured using TEM, and the results are presented in Figure 10A1–A4. The miR-221/222+ group exhibited regenerated nerve fibers with thicker myelin sheaths, which were more similar to the normal nerve fibers (Figure 10B1–B4). The injury site exhibited MBP protein expression during regeneration (Figure 10C1–C4). At 4 weeks post-treatment, very little MBP was detected in the miR-221/222− group (Figure 7C1). However, significantly more MBP expression was observed in the miR-221/222+ group, which showed ring-like positive expression shapes (Figure 10C4). The results of statistical analysis are shown in Figure 10D–F. At 4 weeks posttreatment, the myelin sheath thickness was significantly greater in the miR-221/222+ group compared to the blank group (0.58±0.07 μm vs 0.34±0.09 μm; P<0.05). However, no significant difference between the NC group and the blank group was observed in the thickness of the myelin sheath (0.38±0.06 μm vs 0.34±0.09 μm; P>0.05) (Figure 10D). More unmyelinated axons than myelinated axons were detected in the miR-221/222− group (Figure 10E). Irregular positive spots were seen in the blank and NC group, greater MBP expression than the miR-221/222− group but less than in the miR-221/222+ group (n=6, P<0.05) (Figure 10F).
Discussion
Gene therapy shows great potential in treating enormous amount of illness but lack of an ideal gene delivering system remains one of the major obstacles to application.38,39 In the present study, the presynthesized cationic polymer PDAPEI, which has shown great potential in delivering gene for tumor and rheumatoid arthritis treatment,34,35 was explored for its possible dene delivering application in peripheral nerve regeneration. Mir-221/222 was tested for in vivo nerve regeneration for the first time in our study. PDAPEI/miR-221/222 complexes promoted nerve regeneration after sciatic nerve crush, which may provide a safe therapeutic means of treating nerve crush injury.
PEI25kDa has a higher density of cationic charges, which indicates a greater DNA packing capability and a higher transfection efficiency, but the cytotoxicity limits its wide application.18 A lot of work has been performed to evaluate the cross-linking of low molecular weight PEI, which has been applied in different cell lines. In this study, we synthesized a low molecular weight cross-linked poly (ethylene imine) using PDA and named it PDAPEI. PDAPEI/pDNA shows a low cytotoxicity and relatively high transfection efficiency, with a perfect particle size (60–85 nm) and a constant positive zeta potential (16–21 mV). Furthermore, for all N/P ratios from 1 to 300, no particle size was more than 150 nm (the maximum size for easy endocytosis). Thus, the suitable particle size and zeta potential facilitate crossing the negative-charged cell membrane barrier. The DNA binding capability of nanoparticles is a prerequisite characteristic for protecting gene from degradation in gene delivery systems.40 In the agarose gel electrophoresis assay, the band for pDNA was completely invisible at a N/P ratio of 5 and above, which indicated that the polycationic material PDAPEI can effectively bind the plasmid DNA at a small ratio. In our study, PDAPEI had a relatively low cytotoxicity but supported a higher (80%) cell viability at all N/P ratios tested. We attribute this to the fact that PDAPEI can biodegrade into low molecular weight PEI and 2,6-pyridinedicarboxylic acid in the acidic intracellular environment. Similar degradation process has been validated in our previous study.41 All the characteristics observed demonstrated that PDAPEI is a promising agent for gene delivery. In this study, we achieved a transfection efficiency of 15%, lower than previously nonviral transfection protocol, where a transfection rate of 33% was reported.42 Howbeit a similar transfection efficiency was observed in the positive PEI25kDa control group (12%). Meanwhile, the cell viability was higher than the previous vector in different ratios as follows: more than 90% (N/P: 1–150) and 80% (N/P: 200–300) in our study, demonstrating that PDAPEI is a more biocompatible and lower toxic material. All the experiments were repeated three times, and some of these results were confirmed to be similar to what is reported in the literature.
MicroRNA therapy has been examined in neuroscience and plays an important role in the synaptic plasticity and regeneration of neurons.43–45 SCs participate in the production of NGF and differentiation into myelinating cells after peripheral nerve injury, and miRNA plays an important role in this procedure.46,47 Forty-eight hours after the successful transfection of miR-221/222 into SCs, the expression of NGF and MBP in the miR-221/222+ group was significantly higher than that in the control group. The probable explanation is that miR-221/222 promoted SCs proliferation, and LASS2 is the target.32 Additionally, LASS2 3′-UTR 5′…AAGUUUGGCAUGAUGUAGC… is in alignment with the miR-221 3′-UUUGGGCGU…CUGUUACAUCG, and miR-222 3′-UGGGUCAUCGGU…CUACAUCG, binds to the highly expressed miR-221/222 to promote SCs proliferation and migration.
Lots of miRNAs have been explored for nerve regeneration, but fewer testified in vivo study. MiR-338 and miR-21 were cotransfected for the treatment of 10 mm rat sciatic nerve defect bridged with collagen nerve conduits and got a relatively satisfying therapeutic intervention.28 Similar to this study, we also explored the potential value of the PDAPEI/miR-221/222 complexes for crush nerve injury regeneration in vivo. Myelin is required for the conduction of action potentials and axonal trophic support, which is synthesized by SCs in the peripheral nervous system.48 MiRNAs play an important role in myelinating glia by modulating gene expression patterns in SCs.49–51 The thickness of myelin sheath is a symbol of myelination and maturation. In the animal study, the myelin sheath thickness of the regenerated nerve in miR-221/222+ group was 0.58±0.07 μm, which was much better than that in the control group 0.34±0.09 μm. This suggests that upregulating the expression of miR-221/222 in SCs enhanced myelination. Possible explanation is that the upregulation of miR-221/222 promoted MBP secretion in SCs. These results were confirmed in the miR-221/222− group, where exhibited fewer myelinated nerve fibers and in which MBP expression was not detected. The MBP-positive area was significantly larger in samples from the miR-221/222+ group than in the other three groups, which demonstrates that more myelination occurred in this group. Although the mechanism for miR-221/222 for nerve regeneration is not completely understood yet, our study demonstrated its feasibility and potential value for further study. Meanwhile, our study has a more systemic and comprehensive analysis (both in vitro and in vivo) than pervious miRNA transfection study for peripheral nerve regeneration. Some other treatments have been applied for promotion of regeneration after sciatic nerve crush, such as Sam68, melatonin, and tissue plasminogen.52–55 These studies got a closely approximate functional results as our study. One possible explanation for this phenomenon is that peripheral nerve regeneration is a multicellular complexity problem and multiple factors can affect these processes.56
In this study, PDAPEI was synthesized to deliver miR-221/222 into SCs and showed a lower cytotoxicity and better transfection efficiency and had the same DNA condensation capability compared to PEI25kDa. The upregulated expression of miR-221/222 in SCs enhanced NGF and MBP expression in vitro and remyelination and functional recovery in vivo. Considering the complexity of central nervous system and the limitations of SNCI animal model, all these results were applicable only for peripheral nerve system. The definitive target of PDAPEI/miR-221/222 for peripheral nerve regeneration is not clearly determined, but the results of this study indicate that the use of PDAPEI to deliver miR-221/222 has great potential application in rat sciatic nerve regeneration and is worthy of further study.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Nos 81571261 and 81271999), the Interdisciplinary Projects of Medicine and Engineering of Shanghai JiaoTong University (No YG2015MS06, YG2014QN06 and YG2016QN22), SUMHS seed foundation project (No HMSF-16-21-010), and Natural Science Foundation of Shanghai (15ZR1432500 and 14712400304). The authors would like to thank Wei Zheng and Wei Chen for their help. Jialin Song, Xueyang Li, and Yingli Li share first authorship.
Author contributions
WY conceived the initial idea and the conceptualization, designed the experiments, participated in the data extraction and analysis, and revised the manuscript. JS, XL, and YL conceived and participated in its design, searched databases, extracted, and assessed studies and helped to draft the manuscript. JC, XL, XZ, YC, and XZ participated in the conceptualization and design of the experiment, data extraction, and analysis. JS wrote the manuscript and WY revised the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Burnett MG, Zager EL. Pathophysiology of peripheral nerve injury: a brief review. Neurosurgical Focus. 2004;16(5):E1. | ||
Li BH, Kim SM, Yoo SB, Kim MJ, Jahng JW, Lee JH. Recombinant human nerve growth factor (rhNGF-β) gene transfer promotes regeneration of crush-injured mental nerve in rats. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;113(3):e26–e34. | ||
Song YS, Joe JH, Joo HW, et al. The effects of granulocyte-colony stimulating factor on regeneration in nerve crush injuries in rats. Neurochem Res. 2016;41(7):1645–1450. | ||
Gu X, Ding F, Williams DF. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials. 2014;35(24):6143–6156. | ||
Choudhury SR, Hudry E, Maguire CA, Sena-Esteves M, Breakefield XO, Grandi P. Viral vectors for therapy of neurologic diseases. Neuropharmacology. Epub 2016 Feb 21. | ||
Ramamoorth M, Narvekar A. Non viral vectors in gene therapy – an overview. J Clin Diagn Res. 2015;9(1):GE01–GE06. | ||
Song J, Ouyang Y, Che J, et al. Potential value of miR-221/222 as diagnostic, prognostic and therapeutic biomarkers for diseases. Front Immunol. 2017;8:56. | ||
Ge X, Zhang Q, Cai Y, et al. PEG-PCL-DEX polymersome-protamine vector as an efficient gene delivery system via PEG-guided self-assembly. Nanomedicine (Lond). 2014;9(8):1193–1207. | ||
Ge X, Feng J, Chen S, et al. Biscarbamate cross-linked low molecular weight Polyethylenimine polycation as an efficient intra-cellular delivery cargo for cancer therapy. J Nanobiotech. 2014;12:13. | ||
Chen S, Feng J, Ma L, Liu Z, Yuan W. RNA interference (RNAi) technology for anti-Vascular endothelial growth factor (VEGF) treatment. Expert Opin Drug Deliv. 2014;11(9):1471–1480. | ||
Chen S, Ge X, Chen Y, Lv N, Liu Z, Yuan W. Advance of RNAi in the research of Alzheimer’s disease. Drug Des Dev Ther. 2013; 7:117–125. | ||
Duan S, Ge X, Lu N, Wu F, Yuan W, Jin T. Synthetic polyspermine imidazole-4, 5-amide as an efficient and cytotoxicity-free gene delivery system. Int J Nanomedicine. 2012;7:3813–3822. | ||
Ma L, Wei L, Wu F, Hu Z, Liu Z, Yuan W. Advances with microRNAs in Parkinson’s disease research. Drug Des Dev Ther. 2013;7:1103–1113. | ||
Chen W, Li H, Shi D, Liu Z, Yuan W. Microneedles as a delivery system for gene therapy. Front Pharmacol. 2016,7:137. | ||
Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res. 2012;1:27. | ||
Al-Dosari MS, Xiang G. Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 2009;11(4):671–681. | ||
Zou SM, Erbacher P, Remy JS, Behr JP. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. J Gene Med. 2000;2(2):128–134. | ||
Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60(2):247–266. | ||
Lin G, Hong Z, Huang L. Smart polymeric nanoparticles for cancer gene delivery. Mol Pharm. 2014;12(2):314–321. | ||
Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7(8):992–1009. | ||
Islam MA, Park TE, Singh B, et al. Major degradable polycations as carriers for DNA and siRNA. J Control Release. 2014;193:74–89. | ||
Cho CS. Design and development of degradable polyethylenimines for delivery of DNA and small interfering RNA: an updated review. Int Schol Res Noti. 2012;2012. | ||
Huang X, Shen S, Zhang Z, Zhuang J. Cross-linked polyethylenimine-tripolyphosphate nanoparticles for gene delivery. Int J Nanomedicine. 2014;9:4785–4794. | ||
Chen S, Jin T. Poly-cross-linked PEI through aromatically conjugated imine linkages as a new class of pH-responsive nucleic acids packing cationic polymers. Front Pharmacol. 2016;7:15. | ||
Victor A. The functions of animal microRNAs. J High Energy Physics. 2009;4:689–696. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. | ||
Wang J, Muheremu A, Zhang M, et al. MicroRNA-338 and microRNA-21 co-transfection for the treatment of rat sciatic nerve injury. Neurol Sci. 2016;37(6):883–890. | ||
Yi S, Yuan Y, Chen Q, et al. Regulation of Schwann cell proliferation and migration by miR-1 targeting brain-derived neurotrophic factor after peripheral nerve injury. Sci Rep. 2016;6:29121. | ||
Wu D, Murashov AK. MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front Mol Neurosci. 2013;6:35. | ||
Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276(12):3269–3276. | ||
Yu B, Zhou S, Wang Y, et al. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. J Cell Sci. 2012;125(Pt 11):2675–2683. | ||
Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci. 2012;35(2):123–134. | ||
Song J, Chen Y, Jiang S, et al. Efficient and nontoxic biological response carrier delivering TNF-α shRNA for gene silencing in a murine model of rheumatoid arthritis. Front Immunol. 2016;7:305. | ||
Che J, Tao A, Chen S, Li X, Zhao Y, Yuan W. Biologically responsive carrier-mediated anti-angiogenesis shRNA delivery for tumor treatment. Sci Rep. 2016;6:35661. | ||
Wang CY, Zhang KH, Fan CY, Mo XM, Ruan HJ, Li FF. Aligned natural-synthetic polyblend nanofibers for peripheral nerve regeneration. Acta Biomater. 2011;7(2):634–643. | ||
Thapa L, He CM, Chen H. Study on the expression of angiotensin II (ANG II) receptor subtype 1 (AT1R) in the placenta of pregnancy-induced hypertension. Placenta. 2004;25(7):637–641. | ||
Lee SY, Yang CY, Peng CL, et al. A theranostic micelleplex co-delivering SN-38 and VEGF siRNA for colorectal cancer therapy. Biomaterials. 2016;86:92–105. | ||
Moran G, Feinshtein V, Polyak D, Scomparin A, Satchi-Fainaro R, David A. Inhibition of gene expression and cancer cell migration by CD44v3/6-targeted polyion complexes. Bioconjug Chem. 2016;27(4):947–960. | ||
Zhao D, Gong T, Zhu D, Zhang Z, Sun X. Comprehensive comparison of two new biodegradable gene carriers. Int J Pharm. 2011;413(1–2):260–270. | ||
Xiang S, Su J, Tong H, et al. Biscarbamate cross-linked low molecular weight PEI for delivering IL-1 receptor antagonist gene to synoviocytes for arthritis therapy. Biomaterials. 2012;33(27):6520–6532. | ||
Haastert K, Mauritz C, Chaturvedi S, Grothe C. Human and rat adult Schwann cell cultures: fast and efficient enrichment and highly effective non-viral transfection protocol. Nat Protoc. 2007;2(1):99–104. | ||
Nguyen LH, Diao HJ, Chew SY. MicroRNAs and their potential therapeutic applications in neural tissue engineering. Adv Drug Deliv Rev. 2015;88:53–66. | ||
Svaren J. MicroRNA and transcriptional crosstalk in myelinating glia. Neurochem Int. 2014;77:50–57. | ||
Kaplan BB, Kar AN, Gioio AE, Aschrafi A. MicroRNAs in the axon and presynaptic nerve terminal. Front Cell Neurosci. 2013;7:126–126. | ||
Rodrigo LL, Felipe AC. Schwann cell exosomes mediate neuron–glia communication and enhance axonal regeneration. Cell Mol Neurobiol. 2016;36(3):429–436. | ||
Shi H, Gong Y, Qiang L, et al. Derivation of Schwann cell precursors from neural crest cells resident in bone marrow for cell therapy to improve peripheral nerve regeneration. Biomaterials. 2016;89:25–37. | ||
Nave KA. Myelination and support of axonal integrity by glia. Nature. 2010;468(7321):244–252. | ||
Kang Z, Hong L, Hao H, Mengsheng Q. MicroRNAs and glial cell development. Neuroscientist. 2012;18(2):114–118. | ||
He X, Yu Y, Awatramani R, Lu QR. Unwrapping myelination by microRNAs. Neuroscientist. 2012;18(1):45–55. | ||
Dugas JC, Lucia N. MicroRNAs in oligodendrocyte and Schwann cell differentiation. Dev Neurosci. 2011;33(1):14–20. | ||
Wu W, Liu Y, Wang Y. Sam68 promotes Schwann cell proliferation by enhancing the PI3K/Akt pathway and acts on regeneration after sciatic nerve crush. Biochem Biophys Res Commun. 2016;473(4):1045–1051. | ||
Chang HM, Liu CH, Hsu WM, et al. Proliferative effects of melatonin on Schwann cells: implication for nerve regeneration following peripheral nerve injury. J Pineal Res. 2014;56(3):322–332. | ||
Chang HM, Liu CH, Hsu WM, et al. Proliferative effects of melatonin on Schwann cells: implication for nerve regeneration following peripheral nerve injury. J Pineal Res. 2014;56(3):322–332. | ||
Akassoglou K, Strickland S, Degen JL, Strickland S. Tissue plasminogen activator-mediated fibrinolysis protects against axonal degeneration and demyelination after sciatic nerve injury. J Cell Biol. 2000;149(5):1157–1166. | ||
Cattin AL, Lloyd AC. The multicellular complexity of peripheral nerve regeneration. Curr Opin Neurobiol. 2016;39:38–46. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.