Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 14
Bio-Mechanism Inhibitory Prediction of β-Sitosterol from Kemangi (Ocimum basilicum L.) as an Inhibitor of MurA Enzyme of Oral Bacteria: In vitro and in silico Study
Authors Evangelina IA, Herdiyati Y, Laviana A , Rikmasari R, Zubaedah C, Anisah, Kurnia D
Received 1 February 2021
Accepted for publication 20 April 2021
Published 23 June 2021 Volume 2021:14 Pages 103—115
DOI https://doi.org/10.2147/AABC.S301488
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Juan Fernandez-Recio
Ida Ayu Evangelina,1 Yetty Herdiyati,2 Avi Laviana,1 Rasmi Rikmasari,3 Cucu Zubaedah,4 Anisah,5 Dikdik Kurnia5
1Department of Orthodontics, Faculty of Dentistry – Universitas Padjadjaran, Bandung, Indonesia; 2Department of Pediatric Dentistry, Faculty of Dentistry – Universitas Padjadjaran, Bandung, Indonesia; 3Department of Prosthodontics, Faculty of Dentistry – Universitas Padjadjaran, Bandung, Indonesia; 4Department of Dental Public Health, Faculty of Dentistry – Universitas Padjadjaran, Bandung, Indonesia; 5Department of Chemistry, Faculty of Mathematics and Natural Science – Universitas Padjadjaran, Sumedang, Indonesia
Correspondence: Dikdik Kurnia
Department of Chemistry, Faculty of Mathematics and Natural Science – Universitas Padjadjaran, Jl. Raya Bandung Sumedang km.21, Hegarmanah, Jatinangor, Kabupaten, Sumedang, Jawa Barat, 45363, Indonesia
Tel/Fax +62 22 7794391
Email [email protected]
Background: Dental caries is a widespread disease that causes dental tissue destruction and leads to local and general complications. Gram-positive bacteria including Streptococcus mutans, Streptococcus sanguinis, and Enterococcus faecalis take part in dental caries formation. Gram-positive bacteria have cell walls that consistof a thick layer of peptidoglycan which maintains the strength and rigidity of the bacteria, as well as bacteria guard from internal osmotic pressure. The biosynthesis of peptidoglycan involves many enzymes, including the Mur family, penicillin binding protein (PBP), and sortases.
Purpose: This research has the intention to screen and examine the antibacterial compound of edible plant Kemangi (Ocimum basilicum L.) in terms of how it fights against some oral pathogenic bacteria of E. faecalis ATCC 29212, S. mutans ATCC 25175, and S. sanguinis ATCC 10566.
Materials and Methods: The O. basilicum L. was macerated by several organic solvents to obtain the extracts, before then being purified using several combinations of chromatography methods and the compound was discovered via spectroscopic methods. For the assay against bacteria, the extracts and compounds were tested using agar well diffusion and microdilution assay.
Results: The isolated compound was identified as β-sitosterol. The compound activity against bacteria was evaluated by in vitro assay against S. sanguinis ATCC 10566 and E. faecalis ATCC 29212 with the MIC and MBC value of 25,000 and 50,000 ppm, respectively. The compound was also tested by in silico study using the molecular docking method. The molecular interaction between β-sitosterol and the protein target showed a lower binding affinity value than the native ligand and other positive controls for each protein. Based on the amino acid residue bound to the ligands, β-sitosterol on MurA and SrtA is not competitive to the positive control, showing potential as a natural antibacterial agent. Meanwhile, on the MurB and PBP, β-sitosterol and positive control do compete with each other.
Conclusion: The compound, isolated from O. basilicum L. leaf, was determined as β-sitosterol, which has the molecular formula C29H50O. The antibacterial activity of β-sitosterol by in vitro assay showed weak antibacterial activity, yet exhibited the potential to inhibit the biosynthesis of peptidoglycan and prevent bacteria cell wall formation by inhibiting MurA and SrtA activity via docking simulation.
Keywords: β-sitosterol, Ocimum basilicum L., MurA enzyme, peptidoglycan, cell wall biosynthesis
Introduction
Dental caries is a pathologic process depending on several etiologic factors. The condition causes dental tissue destruction and leads to local and general complications, also being one of the world’s most widespread diseases.1 Acidogenic and aciduric Gram-positive bacteria, such as Streptococcus mutans, Streptococcus sanguinis, and Enterococcus faecalis, was involved in dental caries formation.2,3 Somehow, there is an association between untreated caries and signs of periodontitis. Periodontitis is one of the periodontal diseases which is related to several bacteria such as Porphyromonas gingivalis, Treponema denticola, Fusobacterium nucleatum, and Tannerella forsytiaI. The presence of untreated caries also increases plaque retention and the severity of the periodontal diseases. The association between untreated caries and periodontitis is explained by common risk factors of caries and periodontal diseases, such as the presence of dental biofilm, oral hygiene and living habits, and social factors.4
The cell wall of Gram-positive bacteria is built by a thick layer of peptidoglycan to maintain their strength and rigidity, as well as bacteria guard from internal osmotic pressure.5 Including inhibition of bacterial peptidoglycan formation in cell wall biosynthesis, there are five antibacterial pathways which have already been verified: inhibition of protein synthesis, inhibition of DNA and RNA synthesis, inhibition folate synthesis, and membrane disruption.6 Many kinds of antibiotics and antimicrobial agents are used to prevent dental caries, among which the most widely employed are penicillin, tetracyclines, metronidazole, chlorhexidine, and fluoride.7 Chlorhexidine is the most widely used antiseptic for mouth rinsing and is the gold-standard in dental practice.8 Due to awareness of antibiotics resistance, plant extracts could be an alternative for an antibacterial agent.
The cell wall of bacteria defends bacteria from osmotic pressure and maintains the shape of bacteria.11,36,37 It is primarily composed of peptidoglycan found not only in Gram-positive but also Gram-negative bacteria. Peptidoglycan consists of numerous amino-sugars and short polypeptide chains cross-linked them. The process of enol pyruvate in phosphoenolpyruvate (PEP) being transferred to UDP-N-acetylglucosamine (UNAG) and releasing phosphate in the first step of peptidoglycan biosynthesis was catalyzed by MurA enzyme. As the result, UNAG-enolpyruvate is produced before the reduction is then undertaken, which is catalyzed by the MurB using solvent-derived proton and NADPH. The reduction produces lactyl ether of UDP-MurNAc.38,39
Three main steps occurred in the biosynthesis of peptidoglycan, firstly UDPMurNAc (UDP-N-acetylmuramic acid) formation, secondly addition of short polypeptide chain to UDPMurNAc, and thirdly the unit dispatch through the cytoplasmic membrane and growing peptidoglycan layer.5 UDP-N-acetylglucosamine enolpyruvyl transferase known as MurA and UDP-N-acetylpyruviylglucosamine reductase known as MurB were taking part in peptidoglycan synthesis of the bacterial cell wall at the first step. Both MurA and MurB enzymes catalyzed the formation of UDPMurNAc, the initial precursor of the cell wall biosynthesis.9 Fosfomycin, the antibiotic, specifically modifies and inactive the MurA enzyme.10 The penicillin-binding protein (PBP), existed enzyme in the final step of the bacteria cell wall synthesis and inhibited the transpeptidation reaction, thus leading to the formation of the bacteria cell wall.9,11 Meanwhile, on the cell surface of Gram-positive bacteria, proteins are displayed by sortase enzymes, including sortase A. By linking sortase A to lipid II, proteins are attached on the bacteria cell wall to the cross-bridge peptide.12 For those reasons, The MurA, MurB, PBP, and SrtA are potential targets to develop a new compound with activity against oral pathogenic bacteria.
The Labiatae family is one of the medical plants commonly used not only as spices in food, but also the extracts that have great antibacterial properties. Ocimum is one of the genus from the Labiatae family. It consisted of many species, one of them is O. basilicum L. All the parts from Ocimum plants, as well as extracts and essential oil, were used in various ways, such as spices to give flavor to the food and be effective drugs for many symptoms as told in Africa and Asia folk medicine.13
O. basilicum L. is cultivated in many countries to obtain a regular and maximum supply of the material by natural or greenhouse conditions.14 Besides this, O. basilicum L. extracts are also known for their antibacterial activity. The ethanol extract is reported to have moderate antibacterial activity against S. aureus.15 The activity from extracts of acetone, methanol, and chloroform was evaluated against several bacteria. The methanol extract showed activity against different bacterial strains, such as P. aeruginosa, Shigella sp., L. monocytogenes, S. aureus, and two kinds of E. coli strain. The extracts of acetone and chloroform showed activity against P. aeruginosa bacteria only.13 In previous research, the extracts and active compounds of O. basilicum L. were reported to have antibacterial,13 antioxidant,27 anti-inflammatory,28 antifungal,29 and anticancer benefits.30
Molecular docking is a process involving placing compounds as a ligand in the appropriate configuration to interact with a protein as a receptor. The interaction describes the protein as the main molecular target to easily detect drug action from the compound. The compound or ligand could be obtained from synthesis or isolation from the plant extract process and would bind to the protein target to determine the allosteric or inhibitory effects.25
This research was focusing on determining the active constituent of O. basilicum L. as a lead compound with activity against oral pathogenic bacteria. The compound is then evaluated to determine its activity against oral pathogenic bacteria, S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 through in vitro and in silico study.
Materials and Methods
Materials
O. basilicum L. leaves were cultivated and collected in Ciwidey, Bandung, West Java, Indonesia by local farmers. The specimen was identified and deposited at the laboratory of Taxonomy, Department of Biology, Faculty of Mathematics and Natural Science, Universitas Padjadjaran. Distilled solvents of n-hexane, methanol, and ethyl acetate were used for the extraction, separation, and purification, while the analytical grade organic solvents were used for spectroscopic analysis from Sigma Aldrich Co. Ltd. (St. Louis, MO, USA) and Merck Co. Ltd. The Silica G 60 (Merck, Darmstadt, Germany) and ODS RP-18 were used for column chromatography, and ODS RP-18 F254S and silica G 60 F254 (Merck) plates were used for thin-layer chromatography, respectively. The spot compounds on TLC were visualized under UV light at 254 and 356 nm together by spraying with 10% H2SO4 in EtOH followed by heating.
Antibacterial test of the selected sample against Enterococcus faecalis ATCC 29212, Streptococcus mutans ATCC 25175, Streptococcus sanguinis ATCC 10566 strains used Mueller Hinton agar and broth as a medium. The positive control for this assay was chlorhexidine.
MurA enzyme (protein data bank ID: 1UAE), MurB enzyme (UniProtKB: Q830P3), PBP (protein data bank ID: 6MKG), and SrtA (protein data bank ID: 2KW8) were used for screening antibacterial activity in this research. The 3D structure was retrieved from RSCB Protein Data Bank (https://www.rcsb.org/) and UniProt knowledgebase (https://www.uniprot.org/). Ligands were used in this research, fosfomycin which has ligand bond on MurA enzyme 1UAE was separated from the macromolecule using PYMOL, glycopeptides (CID 56,928,060) as a positive control for MurB enzyme, penicillin (CID 2349) as a positive control for PBP, curcumin (CID 969,516) as a positive control for SrtA, chlorhexidine (CID 9,552,079) as a positive control for antibacterial activity, and beta-sitosterol (CID 222,284) as a sample. All ligands were retrieved from a compound database called PubChem on the website (https://pubchem.ncbi.nlm.nih.gov/).
Instruments
Determining the structure of the isolated compound was guided by spectroscopic methods of several instruments such as infrared (IR) with FTIR Shimadzu 8400, ultraviolet (UV) by 8452A Diode Array, mass spectrometry (MS) with Water Acquit UPLC type triquadrupole, and NMR (1H-NMR, 13C-NMR, DEPT 135°, HMQC, 1H-1H COSY, HMBC) with JEOL type ECA 500 MHz. UV detector lamps with wavelengths of λmax at 365 and 254 nm were used to illuminate the TLC plates. Antibacterial activity of extracts and compound used paper disks, Laminar airflow, incubator Memmert, autoclave machine HVE-50 Hirayama, anaerobic jar, microplate 96 well, microtubes, micropipettes, and Microplate Reader of Biochrom EZ Read 400 ELISA.
Isolation Compound from Extract of O. basilicum L
A sample of 2.8 kg of fresh O. basilicum L. leaf was extracted with methanol (21 L) using the maceration method for 3×24 hours. The filtrate from the maceration process was evaporated by rotary evaporator at 40°C and 103.21 g of methanol extract were obtained. The methanol extract was partitioned using n-hexane-H2O (8x100 mL n-hexane) to obtain 21.94 g of n-hexane extract, following which ethyl acetate-H2O (5x100 mL) was employed to obtain 13.76 g of ethyl acetate extract and 35.15 g of H2O extract, respectively. For the bioactivity evaluation of antibacterial activity, all extracts were made in a series of concentrations, according to assay protocols.16
The n-hexane extract (15 g) which showed the best result activity against S. mutans ATCC 25175 strain was purified by gradual chromatography using Silica G 60 (0.063–0.200 mm) eluted by n-hexane and ethyl acetate with 5% gradient (v/v). Fractions of F.1–21 were obtained and visualized under UV light at 254 and 356 nm together via spraying with 10% H2SO4 in EtOH, which was followed by heating. For the next step, each fraction from F3–F11 was tested for its antibacterial activity using the agar disk Kirby-Bauer method. Among the fractions, fraction 5 (600 mg) showed the greatest inhibition zone (10.3 mm). Then, fraction 5 was purified by column chromatography on silica G 60 using the normal phase method by 1% gradient of n-hexane and ethyl acetate mixture of solvent. From this step, F.5-(1-30) were obtained and also visualized under UV light at 254 and 356 nm together via spraying with 10% H2SO4 in EtOH, which was followed by heating. Subsequently, F.5–8, F.5–9, and F.5–10 (296.4 mg) were purified by column chromatography on silica G 60 using isocratic elution. The mixture of 98 parts of n-hexane and two parts of ethyl acetate (98:2, v/v) solvent was used for elution. Each 5 mL of solvent was collected as a fraction and 215 fractions were obtained. As a result, 80 mg of the pure compound was obtained from F.5-(8-10)-(153-215) fractions.
Structure Determination of Isolated Compound
Comprehensive analysis data of spectroscopic methods were used to determine the structure of an isolated compound, such as ultraviolet (UV) spectrum, mass spectrometry (MS) spectrum, 1D and 2D-NMR spectrum (1H-NMR, 13C-NMR, DEPT 135°, HMQC, 1H-1H COSY, HMBC), and infrared (IR) spectrum.
Assessment of the Extracts and Isolated Compound of O. basilicum L. Leaf Activity Against Oral Pathogenic Bacteria
Kirby-Bauer disk diffusion method was used to evaluate O. basilicum L. extracts activity against S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 strains. This method is commonly employed to ascertain the activity of the sample against oral pathogenic bacteria. The diameter of the growth area around the paper disk was measured to obtain the inhibition zone of each sample.26 The assay determines the resistance or sensitivity of S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 strains to extracts guided by CLSI protocols (CLSI, 2012).17
Methanol was used to dilute all the samples except for water fraction and positive control (chlorhexidine) was diluted in water. Stock solutions were made for each extract, 75 mg of each extract was diluted in methanol except for water fraction to obtain 5% stock solution. Then, 1, 2, 3, 4, and 5% concentration of all samples, together with 2% concentration of chlorhexidine were made for the assay. Then 20 μL of each sample was impregnated to a 6 mm paper disk and was put on the agar surface. A repeated test is required to obtain a better result.
1 ose of bacteria was grown in 5 mL of broth media to prepare the bacteria. Then, the solution was incubated at 37°C for 24 hours. A microplate reader was used to measure the optical density of the solution at 620 nm after the incubation process. The solution was diluted to reach 0.5 McFarland standard or about 180 CFU/mL in broth media (100 µL). On the surface of agar media, the resulting culture was swabbed and the prepared paper disk was put. Moreover, the samples were incubated at 37°C for 48 hours. A repeated test is required to obtain a better result.
Determination of MIC and MBC of the compound against E. faecalis ATCC 29212, and S. sanguinis ATCC 10566 strains were conducted using the microdilution method in 96-well microplate.18 In Mueller Hinton broth at 37°C, the bacterial cells were pre-cultured under aerobic conditions. In the presence of a compound in several concentrations by serial two-fold dilution, bacterial cells were incubated at 37°C without shaking for 48 hours in the same broth on a microplate, as shown in the procedure used at the Clinical and Laboratory Standards Institute. Then, using a microplate reader at 620 nm, the measurement of solution optical density proceeded. The cells’ MICs were defined as the lowest concentrations where visually the bacterial cells were not observed by OD value as reported previously and given by duplicate assessments. The compound was dissolved in methanol where it was a negative control, and chlorhexidine was used as a positive control. On the surface of the agar, each concentration of the compound solution in the microplate well was spread and incubated at 37°C for 24 hours. After incubation, the liquid of each well on the microplate was measured using a microplate reader at 620 nm. Following this, the MIC value was determined by comparing the absorption value of the sample well (compound plus bacteria) with that of the blank well (bacteria). Furthermore, the liquid of wells was spread on the Mueller Hinton agar and incubated for 48 hours to evaluate MBC, the lowest concentration of the sample required to kill the bacteria.
In silico Characterization of β-Sitosterol
The antibacterial activity of β-sitosterol was predicted by the structure’s canonical SMILE using (http://www.pharmaexpert.ru/passonline/), online program PASS ONLINE. It predicted the activity against bacteria of the compound and other bioactivities of the compound. A Pa (probability of being active) value of more than 0.7 was selected.19
Autodock Vina in the open-source software PyRx 0.8 was used for the ligand-protein docking and virtual screening of antibacterial activity. The β-sitosterol and chlorhexidine were subjects for binding to the MurA, MurB, PBP, and SrtA as protein targets, ligands were free for blind docking. Fosfomycin, the native ligand for the MurA enzyme, was being re-docked to the enzyme. Glycopeptide was a specific subject for the MurB enzyme only, while penicillin was specifically for PBP and curcumin was for SrtA. The conformation was selected based on binding energy, the one with the lowest binding energy score that has a value of root-mean-square deviation less than 1.0 Å was selected.19
The docking results of antibacterial activity were visualized by the PYMOL program and analyzed using the Discovery Studio 2020 Client program. Ligand-residue interaction and docking pose in the 3-dimension molecular picture was shown by the PYMOL program. Subsequently, those interactions were shown in the 3-dimension molecular pictures for best visualization using the Discovery Studio 2020 Client program. Each protein–ligand complex was compared to 3-dimension pictures of the MurA, MurB, PBP, SrtA, which bound ligands on fatty acid sites. The similarity from ligation pose of β-sitosterol, chlorhexidine, and specific positive control for each protein target to another compound that bound ligands on fatty acid sites is being related to β-sitosterol bioactivity by its docking pose.
Results
Extraction of Kemangi (O. basilicum L.) Leaf
In total, 103.21 g of methanol extract was obtained from the maceration process. From the partition process, 21.94 g of n-hexane extract, 13.76 of ethyl acetate extract, and 35.15 of water extract were obtained, respectively.
Antibacterial Activity Assay of Extracts Against S. mutans ATCC 25175, S. sanguinis ATCC 10566 and E. faecalis ATCC 29212
The result of the bioactivity assay of O. basilicum L. extracts against S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 strains are shown in Table 1. According to Gherairia et al,20 the antibacterial activity of the Kirby-Bauer assay was classified by five categories of their inhibition zone, which are very strong activity for ≥30 mm, strong activity for 21–29 mm, moderate activity for 16–20 mm, weak activity for 11–15 mm, and little or no activity for ≤0 mm. The n-hexane extract showed a weak response (9.4 mm) against S. mutans ATCC 25175 strain by the concentration of 5%, yet showed the widest diameter of inhibition zones among the extracts. For antibacterial activity against S. sanguinis ATCC 10566 strain by the extracts, ethyl acetate extract showed a moderate response (16.4 mm) by a concentration of 5% and showed the widest diameter of the inhibition zone among the extracts. Furthermore, the antibacterial activity of the extracts against E. faecalis ATCC 29212 strain showed a weak response (10.4 mm) by 5% of methanol extract.
 |
Table 1 Inhibition Zone (mm) of Each Extract of Kemangi against S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 |
Purification of β-Sitosterol from n-Hexane Extract of Kemangi (Ocimum basilicum L.) Leaf
The compound was isolated from the extract which has activity against the bacteria. The active fractions of n-hexane were separated and purified via the bioactivity guided-separation approach using different phase techniques, normal and reverse phase of column chromatography. Normal phase using a stepwise solvent, while the reverse phase was using an isocratic solvent. The purity of compounds was monitored by a combination of 1D and 2D TLC analysis on the normal phase, respectively.
Structural Determination of Compound
The isolated compound was a white crystal with a molecular mass (m/z) of 413.7 as shown on [M+H]− peak by mass spectroscopy ions and related to the molecular formula of β-sitosterol (C29H50O). The compound showed a maximum wavelength of 202 nm as the result of the maximum wavelength measurement shown in the UV spectrum. The compound showed absorption of hydroxyl (OH) at 3,549 cm−1, CH2 at 2,935 cm−1, CH at 2,867 cm−1, C=C at 1,637 cm−1, and C-O at 1,063 cm−1,21 respectively, in the IR spectrum.
The 1H NMR spectrum of the compound showed chemical shift δH 0.68 and 0.99 ppm of two methyl singlets. The presence of three methyl doublets was shown at δH 0.81, 0.83, and 0.93 ppm; δH 0.84 ppm was a chemical shift for methyl triplet. The compound also showed the presence of three protons corresponding to trisubstituted olefinic bonds by the chemical shift of δH 5.36 ppm. The chemical shift of δH 3.56 ppm corresponded to the H-3 proton of a sterol moiety.22
Twenty-nine carbons were shown in the 13C NMR spectrum. DEPT 135° would classify carbons into four categories, namely CH3, CH2, CH, and QC (quaternary carbon), and showed fifteen peaks for six of the CH3 and nine of the CH. A total of 11 down peaks showed the presence of CH2. The result of DEPT 135° indicated the presence of 26 carbons. The missing three carbons were identified as quaternary carbons. Based on chemical shifts of 13C NMR, δC 140.8 and 121.8 indicated the presence of double bond between carbon atoms in positions 5 and 6 (C5=C6); the presence of β-OH for carbon in position 3 (C3) was identified by chemical shift δC 71.9 ppm. Angular methyl carbons in positions 18 and 19 were identified by signals at δC 36.2 and 19.5 ppm, respectively.23 The spectrum of HMBC showed the interaction between proton and carbon which is distanced by two-to-three bonds. HMBC analysis showed interaction between H-16 and C17,18,20; H-17 and C20; H-20 and C21,22; and H-22 and C17,20,23,24.24 Therefore, based on analysis of various of spectra and data, and compared with other published journals, (3S,8S,9S,10R,13R,14S,17R)-17-[(2R, 5R)-5-ethyl-6-methylheptan-2-yl]-10,13-dimethyl-2,3,4,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol], also known as β-sitosterol, was concluded and is shown in Figure 1.
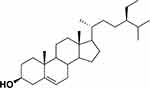 |
Figure 1 Structure of β-sitosterol. |
1H-NMR (CD3OD): δH 3.53 (1H, m, H-3), 5.36 (1H, t, 5.5 Hz, H-5), 0.93 (3H, d, 6.5 Hz, H-19), 0.84 (3H, t, 7.2 Hz, H-24), 0.83 (3H, d, 6.4 Hz, H-26), 0.81 (3H, d, 6.4 Hz, H-27), 0.68 (3H, s, H-28), 0.99 (3H, s, H-29). 13C-NMR (CD3OD): δC 37.3 (C-1), 31.9 (C-2), 71.9 (C-3), 42.3 (C-4), 140.8 (C-5), 121.8 (C-6), 31.7 (C-7), 32.0 (C-8), 50.2 (C-9), 36.6 (C-10), 21.2 (C-11), 39.8 (C-12), 42.4 (C-13), 56.8 (C-14), 26.2 (C-15), 28.3 (C-16), 56.1 (C-17), 36.2 (C-18), 19.5 (C-19), 34.0 (C-20), 26.1 (C-21), 46.0 (C-22), 23.1 (C-23), 12.1 (C-24), 29.2 (C-25), 19.9 (C-26), 19.5 (C-27), 19.1 (C-28), 11.9 (C-29).
Determine Antibacterial Activity (MIC and MBC) of β-Sitosterol
The activity of the compound against bacteria was tested through the assay to obtain the value of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). The result of the assay is illustrated in Table 2. The compound shows the same result for MIC and MBC values against two different bacteria. The result showed that the compound has a MIC value of 25,000 ppm and an MBC value of 50,000 ppm. Compared with the value of the MIC and MBC from chlorhexidine (312.5 ppm of MIC and 625 ppm of MBC for S. sanguinis ATCC 10566 strain, and 3.91 ppm of MIC and 7.8 of MBC for E. faecalis ATCC 29,122 strain) as a positive control, the compound needs a much higher concentration to inhibit and kill the bacteria which means the compound showed a weak activity against S. sanguinis ATCC 10566 and E. faecalis ATCC 29,122 strains.
 |
Table 2 The MIC and MBC Values of β-Sitosterol against S. sanguinis ATCC 10566 and E. faecalis ATCC 29212 |
Prediction of Antibacterial Activity of β-Sitosterol through Molecular Interaction with MurA, MurB, PBP, SrtA
The antibacterial activity of β-sitosterol as an inhibitor for the MurA and MurB enzymes, PBP, and SrtA was determined through molecular docking. Interaction between β-sitosterol and the protein target was evaluated via binding affinity value and intermolecular interaction, such as hydrogen bond. The binding affinity of β-sitosterol (−6.7 kcal/mol) on the MurA enzyme showed a lower value than its native ligand, fosfomycin (−4.2 kcal/mol), and a higher value than chlorhexidine (−8.3 kcal/mol). On the MurB enzyme, β-sitosterol (−7.6 kcal/mol) also showed a lower value of binding affinity than the MurB positive control, glycopeptide (−7.4 kcal/mol), and a higher value than chlorhexidine (−8.7 kcal/mol), which is listed in Table 3. The binding affinity of chlorhexidine on PBP (−9.8 kcal/mol) and SrtA (−6.5 kcal/mol) also showed the lowest value, followed by β-sitosterol (−7.8 kcal/mol for PBP and −6.0 kcal/mol) and positive control for each protein target, namely penicillin (−7.5 kcal/mol) for PBP and curcumin for SrtA (−5.7 kcal/mol), respectively. The interaction among amino acids in the protein target and the ligand is listed in Table 4. Details of the interaction between the ligand and the protein target are shown in Figure 2 for MurA enzyme, Figure 3 for MurB enzyme, Figure 4 for PBP, and Figure 5 for SrtA. The ligands are distinguished by coloring each of them differently; β-sitosterol in purple, chlorhexidine in green, and each positive control for every protein target in orange. The hydrogen bond is shown in green color, while the hydrophobic bond is in purple.
 |
Table 3 Antibacterial Activity Prediction of β-Sitosterol and Positive Controls |
 |
Table 4 List of Hydrogen Bond and Hydrophobic Interaction of β-Sitosterol-Protein Target and Positive Control-Protein Target |
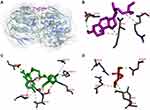 |
Figure 2 Active site of the MurA for: (A) all ligands (B) β-sitosterol, (C) chlorhexidine, and (D) fosfomycin. |
 |
Figure 3 Active site of the MurB for: (A) all ligands (B) β-sitosterol, (C) chlorhexidine, and (D) glycopeptide. |
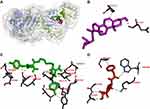 |
Figure 4 Active site of the PBP for: (A) all ligands (B) β-sitosterol, (C) chlorhexidine, and (D) penicillin. |
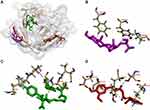 |
Figure 5 Active site of the SrtA for: (A) all ligands (B) β-sitosterol, (C) chlorhexidine, and (D) curcumin. |
Discussion
Discovering active compounds from plants has gained enormous interest from researchers. It involves many disciplines of knowledge including phytochemical, botanical, ethnobotanical, and biological techniques.25 The Labiate family, containing O. basilicum L., specifically the extracts, was known for its activity against bacteria. Therefore, the Kemangi (O. basilicum L.) was selected as a source to obtain new compounds as an antibacterial and compound.13
The antibacterial activity for each extract of O. basilicum L. was tested against S. mutans ATCC 25175, S. sanguinis ATCC 10566, and E. faecalis ATCC 29212 strains by using disk diffusion Kirby-Bauer method to obtain the inhibition zone of each sample. The higher inhibition zone of the extracts showed more effective antibacterial activity against the bacteria. Based on data in Table 1, the n-hexane extract showed the greatest activity against S. mutans ATCC 25175 and for S. sanguinis ATCC 10566 strains, O. basilicum L. extract of ethyl acetate showed the greatest result with inhibition zone 9.4 and 16.4 mm by 5%, respectively. Antibacterial activity of the extracts against E. faecalis ATCC 29212 strain resulted in 10.4 mm of inhibition zone by 5% of methanol extract.
The separation and purification of the O. basilicum L. extract were followed by the result of its activity against S. mutans ATCC 25175 and for S. sanguinis ATCC 10566 strains. Β-sitosterol was obtained from the separation and purification process. β-sitosterol isolation and characterization have been reported in some published journals.21,23,31,32 The structure of β-sitosterol is shown in Figure 1. β-sitosterol is a compound commonly found in several parts of plants producing oil such as grains, fruit, leaves, and stem. It has been reported that β-sitosterol exhibits activity against several bacteria such E. coli and S. aureus, although its activity against S. sanguinis ATCC 10566 and E. faecalis ATCC 29212 strains has not yet been reported.23
Molecular interaction between β-sitosterol as a ligand and a protein as a receptor was also predicted through molecular docking to determine the β-sitosterol mechanism as an antibacterial agent. The scoring function from the docking result is needed to examine the quality of the docking result. Physico-chemical information of geometric complementarity was used to achieve more accurate results, such as binding affinity and interactions that occur between protein and ligand intermolecularly, including hydrogen bond and hydrophobic interaction.25,33 Binding affinity exhibits interaction strengths of a protein and its ligand. It is related to the Gibbs free energy (ΔG).34 It is also used to ascertain the stability bond between a protein and its ligand, as well as to predict the bioactivity value for the complex.35 A donor atom and hydrogen atom are required in one molecule for an intermolecular bond to be formed, as well as an acceptor atom in another molecule. For a hydrogen bond to be formed, the donor atom has to bond to a hydrogen atom, yet the receptor atom is unnecessary to be bonded to a hydrogen atom. The match of the hydrogen atom to its receptor should be one-to-one, but multiple hydrogens could also be matched. The hydrogen bond plays an important role in the structure and its biomolecules function. To find the optimal docked complex, a complex with stronger hydrogen bonds should be chosen, for two arguments. Firstly, the more intermolecular hydrogen bonds exist, the stronger effect for the complex formation, thus the result of the docking will be more precise. Secondly, the displacement of the ligand would be more effective if geometric constraints imposed by the large number of hydrogen bonds were more strict.33
Molecular docking is used to know β-sitosterol antibacterial activity by inhibiting the activity of the MurA and MurB enzymes, also PBP and SrtA, thus meaning that the peptidoglycan biosynthesis is inhibited and the cell wall of bacteria is not formed. Based on the docking result of β-sitosterol and other ligands on different protein targets, the binding affinity value of β-sitosterol is lower than the native ligand or any other positive control, −6.7 for MurA, −7.6 for MurB, −7.8 for PBP, and −6.0 kcal/mol for SrtA, respectively. Chlorhexidine showed the lowest binding affinity than any other ligand, which is the most effective widely-used antiplaque chemical that exhibited great activity against Gram-positive and Gram-negative bacteria and is known as the gold standard.7 Based on binding affinity value, the lower the value, the better the result that will emerge.34 β-sitosterol showed potential as an antibacterial agent by having a lower binding affinity value than other ligands, although it had a higher value than chlorhexidine, the gold standard. Meanwhile, as shown in Figures 2–5, the number of hydrogen bond interactions between receptor and ligand showed that β-sitosterol has a weak complex formation, since it has only one hydrogen bond on Asp51 for MurA, Arg226 for MurB, Glu231 for PBP, and none of it on SrtA, yet it showed many hydrophobic interactions with other residues. This is related to the lack of hydroxyl in the compound. High-affinity ligands require strong hydrogen bonds. Moreover, the protein preferred to act as an H-bonding donor.34 The MurA showed hydrophobic interaction with Lys46 and Arg401 residues. The MurB interacted with Ala138, Ile124, Pro125, Met134, and Ala136 residues. The PBP showed one hydrophobic interaction on the Pro210 residue and SrtA showed four interactions on Tyr149, Pro86, Tyr148, and Tyr150. In addition to the hydrogen bond, hydrophobic interaction is also considered a main driving force behind the conformational change of the protein target.40 Based on Table 4, fosfomycin and chlorhexidine on the MurA enzyme were competitive with each other by binding to the same amino acid residue (Asp305), but not competitive with β-sitosterol, as might be seen in Figure 2. On the MurB enzyme, β-sitosterol is competitive with glycopeptide by binding to the same residue (Pro125), but not competitive with chlorhexidine, although the active sites are close to each other (Figure 3). β-sitosterol and penicillin are competitive with each other on PBP by binding to the same residue (Pro210), but are not competitive with chlorhexidine, despite looking close to the active site of β-sitosterol and chlorhexidine (Figures 4 and 5). Therefore, β-sitosterol will effectively inhibit the biosynthesis of peptidoglycan and prevent cell wall formation by inhibiting the MurA and SrtA activity through having a lower binding affinity than the positive control and not being competitive with the positive control in the active site.
Conclusion
An antibacterial compound has been isolated from n-hexane extract of Kemangi (O. basilicum L.) leaf and showed weak antibacterial activity against S. sanguinis ATCC 10566 and E. faecalis ATCC 29212 strains with the MIC and MBC value of 25,000 and 50,000 ppm, respectively. The isolated compound structure was figured as β-sitosterol, which has the molecular formula C29H50O. The compound shows potential as a blockage of the MurA and SrtA so the biosynthesis of peptidoglycan is inhibited and bacteria cell wall formation is prevented via docking simulation.
Human and Animal Rights
No Animals/Humans were used for studies that are the basis of this research.
Acknowledgments
This research was supported by Academic Leadership Grant 2020 (1427/UN6.3.1/LT/2020 in the name Prof. Yetty Herdiyati) from Universitas Padjadjaran, Sumedang Indonesia. Prof. Yetty Herdiyati acknowledges Universitas Padjadjaran for all research facilities.
Disclosure
The authors reported no conflicts of interest for this work.
References
1. Sicca C, Bobbio E, Quartuccio N, Nicolò G, Cistaro A. Prevention of dental caries: a review of effective treatments. J Clin Exp Dent. 2016;8(5). doi:10.4317/jced.52890
2. Hughes S, Heard K. Antimicrobial therapies for gram-positive infections. Pharm J. 2017;1–22.
3. Palombo EA. Traditional medicinal plant extracts and natural products with activity against oral bacteria: potential application in the prevention and treatment of oral diseases. Evid Based Complement Alternat Med. 2011;2011. doi:10.1093/ecam/nep067
4. Durand R, Roufegarinejad A, Chandad F, et al. Dental caries are positively associated with periodontal disease severity. Clin Oral Investig. 2019;23(10):3811–3819.
5. Smith CA. Structure, function and dynamics in the mur family of bacterial cell wall structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol. 2017;362(2006):640–655. doi:10.1016/j.jmb.2006.07.066
6. Walsh CT, Wencewicz TA. Prospects for new antibiotics: a molecule-centered perspective. J Antibiot (Tokyo). 2014;67:7–22. doi:10.1038/ja.2013.49
7. Qiu W, Zhou Y, Li Z, et al. Application of antibiotics/antimicrobial agents on dental caries. Hindawi. 2020;2020:1–11.
8. Cieplik F, Jakubovics NS, Buchalla W, et al. resistance toward chlorhexidine in oral bacteria – is there cause for concern ? Front Microbiol. 2019;10:587. doi:10.3389/fmicb.2019.00587
9. Nikolaidis I, Favini-Stabile S, Dessen A. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci. 2014;23(3):243–259. doi:10.1002/pro.2414
10. Dai HJ, Parker CN, Bao JJ. Characterization and inhibition study of MurA enzyme by capillary electrophoresis. J Chromatogr B. 2002;766(2001):123–132. doi:10.1016/S0378-4347(01)00461-3
11. Moon TM, D’Andréa ÉD, Lee CW, et al. The structures of penicillin-binding protein 4 (PBP4) and PBP5 from Enterococci provide structural insights into -lactam resistance. J Biol Chem. 2018;293(48):18574–18585. doi:10.1074/jbc.RA118.006052
12. Spirig T, Weiner EM, Clubb RT. Sortase enzymes in gram-positive bacteria. Mol Microbiol. 2013;82(5):1044–1059. doi:10.1111/j.1365-2958.2011.07887.x.Sortase
13. Kaya I, Yigit N, Benli M. Antimicrobial activity of various extracts of Ocimum basilicum l. observation of the inhibition effects on bacteria cells by use of scanning electon microcopy. Complement Altern Med. 2008;5(4):363–369.
14. Jayasinghe CJ, Gotoh N, Aoki T, Wada S. Phenolics composition and antioxidant activity of sweet basil (Ocimum basilicum L.). J Agric Food Chem. 2003;51(15):4442–4449. doi:10.1021/jf034269o
15. Vlase L, Benedec D, Hanganu D, et al. Evaluation of antioxidant and antimicrobial activities and phenolic profile for hyssopus officinalis, Ocimum basilicum and Teucrium chamaedrys. Molecules. 2014;19:5490–5507. doi:10.3390/molecules19055490
16. Wetungu MW, Matasyoh JC, Kinyanjui T, Kinyanjui JC. Antimicrobial activity of solvent extracts from the leaves of Tarchonanthus camphoratus (Asteraceae). J Pharmacogn Phytochem. 2014;3(31):123–127.
17. NCCLS) C and LSI (CLSI-formerly. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard, 11th Ed. Clin Lab Stand Inst; 2012.
18. Rotilie CA, Fass RJ, Prior RB, Perkins RL. Microdilution technique for antimicrobial susceptibility testing of anaerobic bacteria. Antimicrob Agents Chemother. 1975;7(3):311–315. doi:10.1128/aac.7.3.311
19. Suprijono MM, Widjanarko SB, Sujuti H, Kurnia D. Computational study of antioxidant activity and bioavailability of papua red fruit through docking toward human serum albumin. AIP Conf Proc. 2019;2108(1):020020. doi:10.1063/1.5109995
20. Gherairia N, Boukerche S, Chouikh A, Khoudir S, Chefrour A. Antibacterial activity of essential oils from two species of genus thymus growing in different sites of north eastern Algerian. TOM. 2019;XXVI(2):100–104.
21. Saeidnia S. The story of beta-sitosterol - a review. Eur J Med Plants. 2014;4(5):590–609. doi:10.9734/ejmp/2014/7764
22. Chaturvedula VSP, Prakash I. Isolation of stigmasterol and β-sitosterol from the Dichloromethane Extract of Rubus suavissimus. Int Curr Pharm J. 2012;1(9):239–242. doi:10.3329/icpj.v1i9.11613
23. Ododo MM, Choudhury MK, Dekebo AH. Structure elucidation of β-sitosterol with antibacterial activity from the root bark of Malva parviflora. Springerplus. 2016;5(1). doi:10.1186/s40064-016-2894-x
24. Saputra DE, Handayani N, Wartono MW. Isolation and identification of β-sitosterol and stigmasterol mixture from root bark of Slatri (Calophyllum soulattri Burm. f). ALCHEMY J Penelit Kim. 2016;10(1):87. doi:10.20961/alchemy.v10i1.14
25. Hatai B, Banerjee SK. Molecular docking interaction between superoxide dismutase (receptor) and phytochemicals (ligand) from Heliotropium indicum Linn for detection of potential phytoconstituents: new drug design for releasing oxidative stress condition/inflammation of. J Pharmacogn Phytochem. 2019;8(2):1700–1706.
26. Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71–79. doi:10.1016/j.jpha.2015.11.005
27. Kaurinovic B, Popovic M, Trivic S. Antioxidant capacity of Ocimum basilicum L. and Origanum vulgare L. extracts. Molecules. 2011;2011(16):7401–7414. doi:10.3390/molecules16097401
28. Bezerra L, Oliveira A, Pereira B, et al. Anti-inflammatory and antiedematogenic activity of the Ocimum basilicum essential oil and its main compound estragole: in vivo mouse models Jos e. Chem Biol Interact. 2016;257:14–25. doi:10.1016/j.cbi.2016.07.026
29. El-soud NHA, Deabes M, El-kassem LA, Khalil M. Chemical composition and antifungal activity of Ocimum basilicum L. Essential Oil. Open Access Maced J Med Sci. 2015;3(3):374–379.
30. Siddiqui BS, Kabir N, Ahmed S. Anticancer activity of Ocimum basilicum and the effect of ursolic acid on the cytoskeleton of MCF-7 human breast cancer cells. Lett Drug Des Discov. 2010;7(December):726–736. doi:10.2174/1570180811007010726
31. Rahmana SMM, Muktaa ZA, Hossain MA. Isolation and characterization of β-sitosterol-D-glycoside from petroleum extract of the leaves of Ocimum sanctum L.. Asian J Food Agro Ind. 2009;2(January 2009):39–43.
32. Bhat AH, Alia A, Rather GM, Kumar B. Isolation and characterisation of beta-sitosterol from the rhizomes of Arisaema utile and its evaluation for antioxidant activity. Int J Sci Res Biol Sci. 2019;6(2):111–118. doi:10.26438/ijsrbs/v6i2.111118
33. Wu M, Dai D, Yan H. PRL-dock: protein-ligand docking based on hydrogen bond matching and probabilistic relaxation labeling. Proteins. 2012;April:1–17. doi:10.1002/prot.24104
34. Pantsar T, Poso A. Binding affinity via docking: fact and fiction. Molecules. 2018;23(1899):1–11. doi:10.3390/molecules23081899
35. Salomi MV, Aishwarya R, Saranya N. Comparative in silico docking analysis of SOD1 againts natural synthetic antioxidant. Int J Curr Res Acad Rev. 2015;3(1):252–263.
36. Li H, Zhou Y, Wang N, Xin Y, Tang L, Ma Y. Original identification and characterization of a MurA, UDP-N-Acetylglucosamine enolpyruvyl transferase from cariogenic Streptococcus mutans. J Hard Tissue Biol. 2012;21(1):17–24. doi:10.2485/jhtb.21.17
37. Luo H, Liang D, Bao M, et al. In silico identification of potential inhibitors targeting Streptococcus mutans sortase A. Int J Oral Sci. 2017;9(1):53–62. doi:10.1038/ijos.2016.58
38. Nix J, Cunningham ML. Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Acta Crystallogr. 2012;F(68):382–385. doi:10.1107/S1744309112006720
39. Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi:10.1111/j.1574-6976.2008.00104.x
40. Yunta MJR. Docking and ligand binding affinity: uses and pitfalls. Am J Model Optim. 2016;4(3):74–114. doi:10.12691/ajmo-4-3-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
