Back to Journals » OncoTargets and Therapy » Volume 12
BANCR Regulates The Cell Invasion And Migration In Esophageal Squamous Cell Carcinoma Through Wnt/β-Catenin Signaling Pathway
Authors Chen Q, Zheng Y, Wu B, Chen X, Sun F, Ge P, Wang P
Received 14 August 2019
Accepted for publication 24 October 2019
Published 6 November 2019 Volume 2019:12 Pages 9319—9327
DOI https://doi.org/10.2147/OTT.S227220
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Quan Chen, Yiming Zheng, Bingbing Wu, Xia Chen, Fei Sun, Pengfei Ge, Pengcheng Wang
Department of Thoracic Surgery, Taizhou People’s Hospital, Taizhou, Jiangsu Province 225300, People’s Republic of China
Correspondence: Pengcheng Wang
Department of Thoracic Surgery, Taizhou People’s Hospital, No. 366 Taihu Road, Medical High-Tech Zone, Taizhou, Jiangsu Province 225300, People’s Republic of China
Tel/fax +86-13852618808
Email [email protected]
Objective: To explore the regulation of long-chain noncoding BANCR on cell invasion and migration of esophageal squamous carcinoma cells and related mechanisms.
Method: The mRNA expression of BANCR in esophageal squamous carcinoma cells and esophageal squamous cells was detected by quantitative PCR . The relationship between the expression of BANCR and the survival rate of patients with esophageal squamous cell carcinoma (ESCC) was analyzed by Kaplan–Meier method. The BANCR pair was detected by Transwell invasion and scratch test. In ESCC cell lines, the cells had invasion and migration ability; Western blot was applied to detect the expression of proteins involved in the Wnt/β-catenin signaling pathway.
Results: BANCR revealed relatively high expression in esophageal squamous carcinoma cells, and the higher the expression of BANCR was, the lower the survival rate of patients with ESCC was. Inhibition of BANCR expression could effectively reduce the invasion and migration ability of esophageal squamous cell carcinoma. After silencing BANCR, the expression of wnt3a, survivin, β-catenin and c-myc protein was downregulated compared with the negative control group (p<0.05).
Conclusion: Long-chain noncoding BANCR was highly expressed in patients with ESCC and was negatively correlated with patients’ survival time. It was of the capability to modulate the cell migration and invasion of ESCC cells through inducing Wnt/β-catenin signaling pathway.
Keywords: BANCR, ESCC, Wnt/β-catenin, cell migration, cell invasion
Esophageal squamous cell carcinoma (ESCC) is the eighth most cancer worldwide1 and is by far the most common histologic type of esophageal cancer in Asia. Overall, its incidence rates are increasing year by year.2,3 Studies have shown that smoking, drinking, unhealthy diet habit, ethnicity, gene susceptibility and other factors increase the risk of ESCC.4 Despite the development of prevention and diagnostic techniques, patients were mostly diagnosed with ESCC at an advanced stage and still died of recurrence and metastasis for 24 to 48 months after surgery. The survival rate has been below 30% in 5 years.5–7 Therefore, early diagnosis, pathogenesis and prognostic indicators of ESCC are helpful in the prevention and cure of ESCC.
Long-chain noncoding RNAs (lncRNAs) are a class of gene fragments, whose lengths are usually larger than 200 nucleotides and do not have the function of encoding proteins. However, since the discovery of lncRNAs, numerous investigations have revealed that lncRNAs have been participated in life processes by activating or prohibiting cell apoptosis, differentiation, proliferation, invasion and even metastasis.8–13 Just taken lncRNA BCAR4 as an example, it promotes cell migration of breast cancer cells by direct binding with SNIP1 and PNUTS.14 At the same time, overexpression of lncRNA HNF1A-AS1 in lung cancer induces the cell proliferation and metastasis of tumor cells by modulating the protein expression of E-cadherin, cyclin D1 and β-catenin.15 These findings have potentiated lncRNAs’ significant roles in cancer development and progression.
The BANCR was initially discovered by Flockhart et al in 2012 in an RNAsequencing screen. They confirmed that that BANCR was a 693-bp lncRNA with its gene having four exons located on chromosome 9, which regulated the expression of the oncogene BRAFV600E.16 Since then, studies have found that BANCR is significantly elevated in melanin tumor cells and is involved in cell invasion and migration.17 In recent years, the roles of BANCR various human cancers such as non-small-cell lung cancer, papillary thyroid carcinoma, colon cancer have been gradually elucidated.18–20 In 2016, Liu et al found that the induced expression of BANCR promoted tumor development and adverse prognosis of ESCC.21
The crucial Wnt/β-catenin signaling pathway influences tumor development and regulates cell proliferation and differentiation.22 Surging evidences have revealed that the sensitization of Wnt/β-catenin signaling pathway boosts the β-catenin protein’s nuclear translocation, and then activates downstream target genes, leading to ESCC.23 Therefore, we hypothesized that BANCR may affect the cell invasion and multiplication of ESCC cells by modulating the Wnt/β-catenin signaling pathway. In the current study, methods such as Western blot, quantitative PCR (qPCR), Kaplan–Meier analysis, transwell migration assay and scratch adhesion test were combined and used to study the specific expression of BANCR in ESCC cells, influence on cell proliferation and invasion and related molecular mechanisms, which would offer novel approaches for figuring out early diagnosis, treatment and prognosis of ESCC.
Materials And Methods
Materials
Tissue Sample Collection
Eighty pairs (including cancer tissues and adjacent tissues) of ESSC surgical resection specimens ranging from January 2008 to January 2012 were collected from the department of oncology, Taizhou City Hospital. The sample included 45 males and 35 females with ESCC (aged 40−75 years, with a median age of 62 years). This study was confirmed by the medical ethics committee of the hospital, and ESCC in patients was confirmed by the department of histopathology. The excised tumor tissue samples were instantaneously immersed into liquid nitrogen and then transferred to a −80°C freezer for further storage and usage. All patients provided written informed consent before the study, which is in accordance with the Declaration of Helsinki.
Cell Lines And Reagents
Commercial products like the human ESCC cell line KYSE30, TE10 and the human normal esophageal epithelial cell line Het-1A were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. RPMI 1640 medium, fetal calf serum, penicillin and PBS potassium phosphate buffer were purchased from Hyclone, USA. The transfected si-BANCR sequence (GGAAAUAGACUGCAGCACCAATT) and the negative control siRNA (si-NC) sequence (TTCTCCGAACGTGTCACGT) were synthesized by Guangzhou Ruibo Biotechnology Co., Ltd. LipofectamineTM RNAiMAX (Invitrogen, USA) was used as the transfection reagent. Serum-free Opti-MEM medium (Invitrogen) was used for transfection.
Methods
RNA Extraction, Reverse Transcription And Real-Time Fluorescent Quantitative PCR (RT-qPCR)
Total RNAs were extracted from cells and tissues using TRIzol reagent, according to manufcturer's instructions (Invitrogen). The concentration and purity of total RNAs were measured with NanoDrop 2000 and separated by 1% agarose gel with electrophoresis, and RNA at 1.8 to 2.1 A260 nm/A280 was placed at −80°C (RNA has no obvious degradation.). RNA was transcriptionally reversed into cDNA using SYBR Green qPCR SuperMix kit (Invitrogen), according to manufcturer's instructions. The analysis of mRNA relative expression was performed by 2−ΔΔCT.24 The RT-qPCRwas detected by the ABI 7500 System (Applied Biosystems, USA). The primer sequence was as follows: BANCR-F: 5ʹGAAGAGACCCCTGACCCTAA; BANCR-R: 5ʹAAGCCGACTCTCCATACCTT; BANCR-RT: 5ʹGCTCAAACTTGCTCCTGGAT; U6-F:5ʹCTCGCTTCGGCAGCACA; U6-R: 5ʹAACGCTTCACGAATTTGCGT; U6-RT: 5ʹAACGCTTCACGAATTTGCGT.
Cell Culture And Transfection
After cell resuscitation, 10% fetal calf serum and 1% streptomycin were added to RPMI1640 medium to prepare growth medium, and then added to KYSE30, TE10 and Het1A cells. The cells were placed in a volume fraction of 5% CO2 and cultured in an incubator at 37°C. As soon as the cell reached 30% to 50% confluent, the serum-free medium Opti-MEM was applied to replace the growth medium for cell transfection. For transfection, KYSE30 and TE10 cells were treated with 50 nM si-BANCR and si-NC for 4 hrs, respectively, and then replaced with normal growth medium for normal culture.
Scratch Test To Detect Cell Migration Ability
When the cells' growth reached the logarithmic growth phase, they were added into a six-well culture plate at a seeding density of 1 × 106/mL, and cultured in an incubator at a volume fraction of 5% CO2 at 37°C. The cells were observed by microscopy, and after they were grown into a single layer, they were treated with mitomycin for 1 hr to inhibit cell division. Next, a 10-μl sterile micropipette tip was handled to scratch the cell down from the plate. PBS was applied to wash the detached cells, and then the cells were added to serum-free RMPI 1640 medium, and cultured at 37°C in a 5% CO2 incubator. Samples were taken at 0, 6, 24, 48 hrs after scratching, and photographed. Scratch healing was detected by an inverted microscope. Image Pro-Plus 6.0 software was used to randomly measure the scratch width of eight sitesat the same time point of cells. Then, the cell migration rate was calculated, which reflected the cell migration ability of any type or group of ESCC cells. The experiment was repeated thrice. The mobility of the cells was calculated using the following formula: [(1 - scratch time width at other time points)/0 hr scratch width)] × 100% (same sample).
Transwell Assay
Matrigel (BD, USA) was pretreated with a precooled serum-free medium at ratio of 1:3 (V/V), and then 40 µL of the mixture was added to the cold Transwell chamber. The mixture was incubated at 37°C for 2 hrs to obtain a solidified Matrigel. The residual liquid left was aspirated, and 100 µL, 600 µL of serum-free medium was added to both chambers including the upper and lower, and equilibrated at 37°C overnight. The transfected cells were added into the upper chamber at a cell density of 1 × 105/mL, 600 µL of growth medium was then added into the lower chamber, and incubated at 37°C in a 5% CO2 incubator. After 24 and 48 hrs of incubation, the chamber was withdrawn and the cells in the upper chamber were cleaned with a cotton swab and then 4% paraformaldehyde was applied to fix the cells for 15 min. After the cells were once washed with PBS, crystal violet was applied for 10 min to stain the cells, and again the cells were washed with PBS. Then, the cells were observed under an inverted microscope to check if the cells have gone through the small holes and photographed. The experiment was performed thrice.
Western Blot Detection Of Protein Levels
The cells were lysed with RIPA extraction by initially shaking for 15 min. After centrifugation, the supernatant was drawn out, and the protein concentration was determined by bicinchoninic acid assay. The lysate was mixed with gel loading buffer at a volume ratio of 1:1 and then denatured at 100°C for 8 min. Then, 20 µg of proteins was loaded onto SDS-PAGE for separation. After separation, the proteins were transferred onto polyvinylidene difluoride (PVDF) (Millipore Co, USA) membranes. After transferring, the membranes were blocked with 5% nonfat milk diluted with TBST (10 mmol/L Tris-HCl, 0.1% Tween 20, 150 mmol/L NaCl) at room temperature for 2 hrs. After blocking, the PVDF membranes were washed thrice with TBST and then incubated with corresponding primary antibody, including anti-β-catenin antibody (ab16051, Abcam), anti-c-Myc antibody (ab32072, Abcam), anti-wnt3a (ab28472, abcam), anti-survivin (Ab469, abcam) and anti-GAPDH antibody (ab181602, Abcam) at 4°C overnight. On the second day, the membranes (horseradish peroxidase-labeled goat anti-rabbit IgG)were thoroughly washed with TBST, and then diluted with TBST buffer at room temperature for 2 hrs. After washing, the enhanced chemiluminescence reagent was subjected to fluorescent color development, and imaging system was applied to detect the signal and analyze the relative expressions of each protein.
Statistical Analysis
SPSS 16.0 software was used to conduct statistical analysis. The data were written as x ± s, t-test was used to analyze the measurement data, and chi-square test was applied to analyze the count data. At the same time, the data of survival rate of patients with ESCC were analyzed by Kaplan–Meier survival curve and log-rank test. The follow-up period was January 2018. The survival time was defined as the duration from initial treatment to pass-by. Graphs are plotted using GraphPad Prism 6.0, and p<0.05 was taken as the statistical difference.
Results
Expression Of BANCR Gene In ESCC Tissues And Cell Lines
The gene expressions of BANCR in both ESCC cell lines and tissues were detected using RT-PCR. Figure 1A shows the relative expression of BANCR in ESCC tissues was significantly higher than that in adjacent tissues (6.64+0.73, p<0.05). Furthermore, Figure 1B reveals that the expression of BANCR in ESCC cell lines KYSE30 and TE10 was higher than that in normal control group Het-1A (p<0.05), and the growth rate, invasion and migration ability of these two kinds of ESCC cells were similar, so the two kinds of cells were selected for the next experiment.
Correlation Between BANCR And Clinicopathological Characteristics Of Patients With ESCC
The clinical-pathological parameters of 80 subjects with ESCC were analyzed. The median (5.92) of BANCR relative expression was cut into two groups, including either low or high expression group. The data failed to reveal significant difference in BANCR between patients of different ages and genders (p>0.05). The expression of BANCR was related to the pathological stage of ESCC and lymph node metastasis. The higher the pathological stage, the expression of BANCR. The higher the rate (p<0.05), and in patients with lymph node metastasis, the expression of BANCR was also significantly higher than that of patients without metastasis (p<0.01) (Table 1).
 |
Table 1 Relationship Between Expression Of BANCR And Clinicopathological Features Of ESCC Patients |
The Correlation Between BANCR Level And The Medical Diagnosis Of Patients With ESCC
The correlation between the BANCR level and the prognosis survival time of ESCC patients was analyzed by the log-rank test and Kaplan–Meier survival curve. As shown in Figure 2, the survival time of patients with high expression of BANCR was shorter than that of patients with low expression of BANCR (p<0.05). The median survival time of patients with low expression of BANCR and high expression of BANCR was 34 months and 18 months, respectively. The risk of postoperative death was 2.07 times higher than patients in the group of low BANCR expression. All of these data indicated the positive correlation between the expression of BANCR and the severity of patients’ survival with ESCC.
 |
Figure 2 Cumulative survival rates of patients with high and low BANCR expression levels. |
Effect Of BANCR On Migration Ability Of KYSE30 And T10 Cells
After KYSE30 and T10 cells were transfected with siRNA-BANCR, the change in cell mobility from 0 hr to 48 hrs after inhibition of BANCR expression was detected by a scratch test. The results indicated that the cell migration of the two cells significantly reduced after 24 hrs of inhibition of BANCR expression (p<0.01), and the mobility of the cells with siRNA-BANCR treatment was also much lower after 48 hrs than the NC treatment (p<0.05) (Figures 3 and 5A). The above experimental results indicated that inhibition of BANCR expression could effectively attenuate the migration ability of KYSE30 and T10 cells.
 |
Figure 3 The effects of BANCR on migration of KYSE30 and TE10 cells. |
Effect Of BANCR On The Invasion Ability Of KYSE30 And TE10 Cells
Transwell assay were applied to examine the effects of BANCR on cell invasive ability of KYSE30 and TE10 cells. It indicated that the number of cells invaded by the siRNA-BANCR treatment was much lower (p<0.05) when the two types of cell line were transfected with siRNA-BANCR (Figures 4 and 5B). This indicated that the inhibition of BANCR expression significantly attenuates the invasive ability of KYSE30 cells.
 |
Figure 4 The effects of BANCR on invasion of KYSE30 and TE10 cells. |
Effect Of BANCR On Related Proteins In Wnt/β-Catenin Signaling Pathway
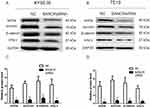
The assay of protein expressions suggested that compared with NC group, silencing of BANCR in both KYSE30 and TE10 cells significantly suppressed the protein expressions of wnt3a, c-myc protein, survivin and β-catenin (p<0.05, Figure 6). The results indicated that downregulation of BANCR affected the proteins involved in the signaling pathway of Wnt/β-catenin.
Discussion
ESCC remains one of the most common types of esophageal cancer, and its early diagnosis, pathogenesis and prognostic indicators are of great significance. In this paper, RT-qPCR, Kaplan–Meier analysis, Transwell experiment, scratch test and Western blot showed that the expression of BANCR gene was relatively high in tumor tissues of ESCC patients, and the higher expression of BANCR indicated the worse of the survival rate of patients with ESCC. Inhibition of BANCR expression could effectively reduce the cell invasion and migration of ESCC, and the protein levels of four important proteins (wnt3a, survivin, β-catenin and c-myc) in the Wnt/β-catenin signaling pathway were significantly decreased. The study suggested that BANCR induced the cell migration and invasion of esophageal squamous carcinoma cells by promoting the Wnt/β-catenin signaling pathway.
Although lncRNA is a kind of RNA molecule that does not have the function of encoding protein, with the technological advance of gene sequencing, its role in gene regulation has attracted wide attention.25 BANCR exhibits high expression in gastric cancer as a lncRNA, and promoted cell proliferation and migration of cancer cells by modulating NF-κB signaling pathway.26 Li et al have found BANCR is highly expressed in human malignant melanoma cell lines and tissues, and silencing of BANCR inhibited cell proliferation by inhibiting MAPK signaling pathway.17 At the same time, similar conclusions have been made in both colorectal cancer and papillary thyroid carcinoma.19 This suggests that BANCR may be highly expressed in numerous types of tumor tissues and revealed a carcinogenic role in the development of tumors. Therefore, we tested BANCR expression levels by RT-PCR in ESCC tissues and cells to demonstrate high expression of the BANCR gene in ESCC. At the same time, we further explained that BANCR was closely related to the prognosis of ESCC by Kaplan–Meier analysis. The higher the expression of BANCR was, the shorter the survival time of ESCC patients was. This result further supported the results of Liu et al.21 In addition, we also used the scratch test and Transwell test to elucidate the regulation of BANCR on ESCC cell migration and invasion. It suggested that the expression of BANCR promoted the cell migration and invasion capacity of ESCC cells.
The Wnt/β-catenin signaling pathway significantly contributes to the development of human cancer.26–30 He et al show that baicalein can target c-myc gene through Wnt/β-catenin signaling pathway to inhibit osteosarcoma cell proliferation and promote cell apoptosis in which jnk, bancr and β-catenin are also involved.31 Xiang et al have indicated that the overexpression of lncRNA DGCR5 can repress hepatocellular carcinoma progression through inhibiting Wnt/β-catenin signaling pathway.32 In recent years, researches have reported that the Wnt/β-catenin signaling pathway played vital roles in the development and progress of ESCC, such as proliferation, invasion, migration and apoptosis.33 Wnt/β-catenin signaling pathway is one of the most important signaling pathways to induce cancer epithelial-mesenchymal transition process which can promote the invasion and migration of cancer cells.34 Li et al have explored that cir-ITCH inhibits the activation of Wnt/β-catenin signaling pathway in ESCC by degrading Dvl2 phosphorylation.35 It has been reported that WNT5A can inhibit the downstream regulations of Wnt/β-catenin signaling pathway by suppressing both the protein expression of β-catenin and its transcriptional activity in ESCC cells.36 There is a study suggested that enhanced Capn4 expression activates the Wnt/β-catenin signaling pathway, resulting in increased ZEB1 expression and the promotion of ESCC cell metastasis.37 The results of protein expressions indicate that silencing of BANCR in both KYSE30 and TE10 cells significantly suppressed the protein expressions of wnt3a, c-myc protein, survivin and β-catenin. Therefore, we believe that BANCR promotes cell migration and invasion of ESCC cells by affecting the Wnt/β-catenin signaling pathway.
In summary, long-chain noncoding BANCR was highly expressed in individuals withESCC, and the higher the expression was, the shorter the survival of ESCC patients was. At the same time, it could also regulate the cell migration and invasion of ESCC cells via activating the signal of Wnt/β-catenin. This study reveals promising novel strategies and specific targets for the pre-diagnosis, treatment and prognosis analysis of ESCC.
Abbreviations
qPCR, quantitative PCR; ESCC, esophageal squamous cell carcinoma; lncRNAs, long-chain noncoding RNAs; NC, negative control.
Acknowledgment
The authors acknowledge the Research Fund Project of Taizhou People’s Hospital (ZL201934).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.v61:2
2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi:10.1002/ijc.25516
3. Yang CS, Chen X, Tu S. Etiology and prevention of esophageal cancer. Gastrointest Tumors. 2016;3(1):3–16. doi:10.1159/000443155
4. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
5. Hu N, Wang Z, Song X, et al. Genome-wide association study of gastric adenocarcinoma in Asia: a comparison of associations between cardia and non-cardia tumours. Gut. 2016;65(10):1611–1618. doi:10.1136/gutjnl-2015-309340
6. Xu Y, Zheng Y, Sun X, et al. Concurrent radiotherapy with gefitinib in elderly patients with esophageal squamous cell carcinoma: preliminary results of a phase II study. Oncotarget. 2015;6(35):38429–38439. doi:10.18632/oncotarget.v6i35
7. He LR, Liu MZ, Li BK, et al. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer. 2010;127(1):138–147. doi:10.1002/ijc.25031
8. Chen Y, Xie H, Zou Y, et al. Tetracycline-controllable artificial microRNA-HOTAIR + EZH2 suppressed the progression of bladder cancer cells. Mol Biosyst. 2017;13(8):1597–1607. doi:10.1039/C7MB00202E
9. Li J, Zhuang C, Liu Y, et al. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin Cancer Res. 2016;35(1):99. doi:10.1186/s13046-016-0372-5
10. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
11. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
12. Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–56218. doi:10.18632/oncotarget.v7i35
13. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi:10.1038/ng.3192
14. Xing Z, Lin A, Li C, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–1125. doi:10.1016/j.cell.2014.10.013
15. Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. doi:10.18632/oncotarget.3247
16. Flockhart RJ, Webster DE, Qu K, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–1014. doi:10.1101/gr.140061.112
17. Li R, Zhang L, Jia L, et al. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9(6):e100893. doi:10.1371/journal.pone.0100893
18. Guo Q, Zhao Y, Chen J, et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8(2):869–875. doi:10.3892/ol.2014.2154
19. Wang Y, Guo Q, Zhao Y, et al. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014;8(5):1947–1952. doi:10.3892/ol.2014.2487
20. Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi:10.1186/1476-4598-13-68
21. Liu Z, Yang T, Xu Z, Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;82:406–412. doi:10.1016/j.biopha.2016.05.014
22. Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19(2):150–158. doi:10.1016/j.ceb.2007.02.007
23. Wang JH, Yuan LJ, Liang RX, et al. GOLPH3 promotes cell proliferation and tumorigenicity in esophageal squamous cell carcinoma via mTOR and Wnt/beta-catenin signal activation. Mol Med Rep. 2017;16(5):7138–7144. doi:10.3892/mmr.2017.7495
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
25. Yao W, Bai Y, Li Y, et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37(4):4305–4312. doi:10.1007/s13277-015-4223-3
26. Zhang ZX, Liu ZQ, Jiang B, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem Biophys Res Commun. 2015;465(2):225–231. doi:10.1016/j.bbrc.2015.07.158
27. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi:10.1016/j.cell.2006.10.018
28. Lu W, Lin C, Li Y. Rottlerin induces Wnt co-receptor LRP6 degradation and suppresses both Wnt/beta-catenin and mTORC1 signaling in prostate and breast cancer cells. Cell Signal. 2014;26(6):1303–1309. doi:10.1016/j.cellsig.2014.02.018
29. White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142(2):219–232. doi:10.1053/j.gastro.2011.12.001
30. Teng Y, Wang X, Wang Y, Ma D. Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Biophys Res Commun. 2010;392(3):373–379. doi:10.1016/j.bbrc.2010.01.028
31. He N, Zhang Z. Baicalein suppresses the viability of MG-63 osteosarcoma cells through inhibiting c-MYC expression via Wnt signaling pathway. Mol Cell Biochem. 2015;405(1–2):187–196. doi:10.1007/s11010-015-2410-6
32. Wang XL, Shi M, Xiang T, Bu YZ. Long noncoding RNA DGCR5 represses hepatocellular carcinoma progression by inactivating Wnt signaling pathway. J Cell Biochem. 2019;120(1):275–282. doi:10.1002/jcb.27342
33. Macdonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi:10.1016/j.devcel.2009.06.016
34. Garg M. Epithelial-mesenchymal transition - activating transcription factors - multifunctional regulators in cancer. World J Stem Cells. 2013;5(4):188–195. doi:10.4252/wjsc.v5.i4.188
35. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi:10.18632/oncotarget.3469
36. Li J, Ying J, Fan Y, et al. WNT5A antagonizes WNT/beta-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol Ther. 2010;10(6):617–624. doi:10.4161/cbt.10.6.12609
37. Zhao YL, Li JB, Li YJ, Li SJ, Zhou SH, Xia H. Capn4 promotes esophageal squamous cell carcinoma metastasis by regulating ZEB1 through the Wnt/beta-catenin signaling pathway. Thorac Cancer. 2019;10(1):24–32.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.