Back to Journals » Infection and Drug Resistance » Volume 16
Bacterial Epidemiology and Antimicrobial Resistance Profiles of Respiratory Specimens of Children with Pneumonia in Hainan, China
Authors Mai W, Liu Y, Meng Q, Xu J , Wu J
Received 14 November 2022
Accepted for publication 2 January 2023
Published 12 January 2023 Volume 2023:16 Pages 249—261
DOI https://doi.org/10.2147/IDR.S397513
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wenhui Mai,1,2,* Yiwei Liu,1,3,* Qiaoyi Meng,1,3 Jianping Xu,1,4 Jinyan Wu1,3,5
1Center for Science Experiments, Hainan Medical University, Haikou, Hainan Province, People’s Republic of China; 2Laboratory Department, Haikou Maternal and Child Health Hospital, Haikou, Hainan Province, People’s Republic of China; 3Department of Pathogen Biology, Hainan Medical University, Haikou, Hainan Province, People’s Republic of China; 4Department of Biology, McMaster University, Hamilton, Ontario, Canada; 5Key Laboratory of Tropical Translational Medicine of Ministry of Education, Hainan Medical University, Haikou, Hainan Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianping Xu; Jinyan Wu, Email [email protected]; [email protected]
Purpose: To investigate the bacterial species and antimicrobial susceptibility of respiratory specimens of children with pneumonia in Hainan, China.
Methods: A total of 5017 specimens, including 4986 sputum samples, 19 bronchoalveolar lavage fluid samples and 12 tracheal tube tip samples from hospitalized children with pneumonia from April 1, 2021 to March 31, 2022 were studied. All the bacterial isolates were identified and confirmed with the VITEK 2 system. Antimicrobial susceptibility of all isolates was determined using the Kirby-Bauer method or the VITEK 2 Compact automatic system, following the breakpoints recommended by the Clinical and Laboratory Standards Institute.
Results: A total of 996 bacterial isolates were collected and classified into 24 species. The top 10 most frequent species were Haemophilus influenzae (356 isolates, 35.7%), Streptococcus pneumoniae (128, 12.9%), Moraxella catarrhalis (114, 11.5%), Escherichia coli (89, 8.9%), Staphylococcus aureus (89, 8.9%), Klebsiella pneumoniae (82, 8.2%), Acinetobacter baumannii (31, 3.1%), Pseudomonas aeruginosa (28, 2.8%), Enterobacter cloacae (18, 1.8%), and Streptococcus agalactiae (13, 1.3%). 70.5% strains had the resistant (R) and/or intermediate (I) phenotypes to at least one of the tested drugs, with a large proportion (54.6%) showing resistance to two or more commonly used antibiotics. In addition, 60.5% (69/114) of M. catarrhalis strains and 42.9% (153/356) of H. influenzae strains produced β-lactamases while 19.1% (17/89) E. coli and 6.1% (5/82) K. pneumoniae strains produced extended-spectrum β-lactamases.
Conclusion: A diversity of pathogenic bacteria were isolated from the respiratory tract of children with pneumonia in Hainan, China. High-frequency resistance to first-line antimicrobial drugs was observed in Gram-negative and Gram-positive bacteria, including 544 isolates resistant to at least two antibiotics. Rapid identification and susceptibility testing should be implemented for children with bacterial pneumonia in Hainan before drug treatment is recommended.
Keywords: pneumonia, bacteria, antimicrobial resistance, children, multidrug-resistant bacteria
Introduction
Pneumonia is one of the major causes of childhood morbidity and mortality globally.1 In 2015, an estimated 921,000 children younger than 5 years old died of pneumonia in the world.2 In China, due to general improvements in living conditions, better healthcare and improved vaccines, the estimates of clinical pneumonia in children decreased from 25.9% in 2000 to 8.4% in 2015.1,3,4 However, an estimated >700,000 children are still hospitalized with pneumonia in China each year, representing a huge disease burden.1,5 These hospitalized pneumonia cases are a major cause of morbidity in children.1,6
Bacteria such as H. influenzae, S. pneumoniae are common causes of pneumonia in children. Based on the 2015 Global Burden of Diseases (GBD) data approximately 64% of pneumonia deaths in children under 5-years old were due to bacterial infections.1,2 For example, through direct transthoracic needle lung biopsy and autopsy, bacteria were found to be responsible for most pneumonia deaths in African children.7 More recently, bacteria were also recognized as causes for secondary pulmonary infections associated with coronavirus disease (COVID-19), increasing morbidity and mortality of COVID patients.8
Antimicrobial resistance (AMR) is recognized as one of the most serious global threats to human health and is commonly associated with high morbidity and mortality rates and large medical cost burdens.9,10 The World Health Organization (WHO) has highlighted that infections by antibiotic-resistant bacteria are responsible for around 700,000 deaths per year worldwide, if left to persist at the current rate, may lead to 10 million deaths in 2050.11 In particular, several recent reports described increases in multidrug resistant (MDR) bacteria during the COVID-19 pandemic, with the rise in MDR bacteria rendering infections increasingly difficult to treat.12–23 Common MDR bacteria include Vancomycin resistant Enterococci (VRE), methicillin (oxacillin) resistant S. aureus (MRSA) and certain Gram-negatives such as E. coli and K. pneumoniae producing extended-spectrum β-lactamases or carbapenem-resistant Enterobacteriaceae (CRE).24,25 Options for treating patients with MDR bacterial infections are often extremely limited.
To effectively prevent and treat infections by MDR bacteria, including those causing pneumonia, we need accurate diagnosis of both the disease agents and their antimicrobial susceptibilities. However, at present, relatively little is known about the bacterial species and antimicrobial susceptibility of respiratory specimens from children with pneumonia, especially in tropical and developing regions such as in Hainan Island, South China. In this study, we aim to investigate the bacterial species and their susceptibilities to commonly used antimicrobial drugs of respiratory specimens from children with pneumonia in Haikou Maternal and Child Health Hospital in Hainan, China. The prevalence of MDR bacteria was also investigated in this study.
Materials and Methods
Samples
A total of 5017 specimens including 4986 sputum samples, 19 bronchoalveolar lavage fluid samples and 12 tracheal tube tip samples from hospitalized children with pneumonia from April 1, 2021, to March 31, 2022, were collected, following standard clinical specimen collection protocols. Repeated strains of the same patient at the same hospitalization episode were excluded. The sputum samples were taken by a disposable sterile suction tube under negative pressure and sent for microscopic examination within 24 hours after admission. Sputum quality was considered adequate if it contained ≥25 leukocytes and ≤10 epithelial cells under low magnification. The Ethics Committee of Hainan Maternal and Child Health Hospital and Hainan Medical University approved our sampling and study. This study did not involve any medical intervention of any patients. The guardians of all patients were provided full information about this study and all of them signed a consent form. The study cohort consisted of 2994 males and 2023 females, of whom 1638 patients were less than 1 month old, 1762 patients were 1 month to 1 year old, 1403 patients were 2 to 5 years old, and 214 patients were 6 to 14 years old (Table S1).
Identification of Bacterial Species
To identify the bacteria, we plated the specimens on Columbia blood plate and chocolate plate.25 The plates were cultured overnight at 35°C in a 5% CO2 incubator. Actively growing and morphologically distinct colonies on each plate were identified to the species level using the VITEK 2 Compact microbial identification system (BioMérieux, France). The strains were stored in a −80°C freezer until being revived for antimicrobial susceptibility testing.
Antimicrobial Susceptibility Testing (AST)
For each bacterial isolate that we obtained from each of the patients, we determined its susceptibility to multiple selected antimicrobial agents. These selected agents differed among species with isolates from each species tested for their susceptibilities to a panel of the most commonly used antimicrobial drugs in China for treating the respective infections caused by each bacterial species. We used the Kirby-Bauer (KB) disk diffusion method to determine the antimicrobial susceptibilities of strains of H. influenzae and M. catarrhalis. For strains of other bacterial species, we used the VITEK 2 Compact automatic system with the AST-GN/GP cards (bioMérieux, France). The specific antibiotics tested for each species are shown in Tables 1 and 2. For the KB disk diffusion method, the disks (Oxoid Ltd., Basingstoke, UK) were placed onto Mueller–Hinton agar (Oxoid Ltd., Basingstoke, UK) in accordance with the Clinical and Laboratory Standards Institute (CLSI) 2021 guidelines.26 H. influenzae ATCC49247 and S. aureus ATCC29213 were used for quality controls. CRE strains were defined as strains of Enterobacteriaceae that were resistant to either ertapenem and/or imipenem.
 |
Table 1 Antibiotic Resistance Rates of Gram-Positive Bacteria Analyzed in This Study |
 |
Table 2 Antibiotic Resistance Rates of Gram-Negative Bacteria Analyzed in This Study |
β-Lactamase Test
The nitrocefin discs were used to detect β-lactamase activity, following established protocol.26 A positive result was defined as a disk color change from white to red. A negative result was defined as no color change. Strains S. aureus ATCC29213 and S. aureus ATCC25923 were used for positive and negative controls.
Extended-Spectrum β-Lactamase Test
For certain groups of bacteria, their ability to produce extended-spectrum β-lactamases was determined by means of a three-disk synergy test (with cefotaxime and cefotaxime/clavulanic acid, ceftazidime, and ceftazidime/clavulanic acid, cefepime and cefepime/clavulanic acid). Strains positive for the production of extended-spectrum β-lactamases were those that showed a greater than or equal to eightfold decrease in MICs for cefotaxime or ceftazidime when tested in combination with clavulanic acid, when compared with their MICs without clavulanic acid, following the Clinical and Laboratory Standards Institute (CLSI) 2021 guidelines.26
Analyses of Data
All statistical comparisons among samples were conducted in GraphPad Prism 8 using the chi-square test. P value <0.05 was considered statistically significant.
Results
Among the 5017 respiratory tract specimens from hospitalized children with pneumonia, we successfully isolated 996 bacterial strains, yielding an overall 19.9% isolation rate for bacteria. Table S1 summarizes the bacteria carriage rates in the respiratory tract among host groups from Haikou Maternal and Child Health Hospital, of different sexes, ages, and different seasons. Our analyses revealed no statistically significant difference in bacteria isolation rates between the two sexes (P = 0.402). However, significant differences were found among the age groups (less than 1 month old, 1 month to 1 year old, 2 to 5 years old, and 6 to 14 years old) (P = 0.000); and among the four seasons (spring, summer, autumn and winter) (P = 0.000). Specifically, patients aged 1 month to 1 year old had the highest bacteria carriage rate (29.5%), followed by those aged 2 to 5 years old (22.7%), and those aged 6 to 14 years old (9.8%), patients aged less than 1 month old had the lowest bacteria carriage rate (8.4%). Pairwise age-group comparisons revealed that except between the <1 month old and the 6 to 14 years old (P > 0.05), all other age groups comparisons showed statistically significant differences in their bacterial carriage rates (P < 0.05). Among the four seasons, the winter samples had the highest bacterial carriage rate (26.2%), the summer samples had the lowest bacterial carriage rate (16.7%), and the other two seasons (spring and autumn) were in-between, at 20.4% and 18.4%, respectively.
We found no statistically significant difference in species composition between bacteria samples from the two sex groups (P = 0.949) (Figure 1). However, bacteria populations from the four age groups (Figure 2) and four seasons (Figure 3) differed significantly in their bacteria species distributions (P < 0.01 in both comparisons). Specifically, the samples from the 2 to 5 years old analyzed here had a significantly higher percentage of H. influenzae (55.2%) than those from less than 1 month (3.6%). In contrast, samples from less than 1 month had a significantly higher percentage of E. coli (27.0%) than those from 2 to 5 years old (0.3%) and 6 to 14 years old (0.0%) (Figure 2). The samples from spring and winter had the higher percentage of H. influenzae (45.0% and 47.4%) than those from summer and autumn (25.6% and 24.4%) (Figure 3).
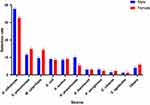 |
Figure 1 Detection rate of respiratory tract bacteria by gender in children with pneumonia. |
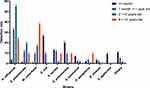 |
Figure 2 Detection rate of respiratory tract bacteria in different age groups. |
 |
Figure 3 Detection rate of respiratory tract bacteria in different seasons. |
Bacteria Isolates
The 996 bacteria isolates belonged to 24 species. Among the 996 isolates, 234 were Gram-positive bacteria (23.5%) and 762 were Gram-negative bacteria (76.5%). Based on their overall frequencies, from the most common to the least common in our sample, the 24 species were H. influenzae (356 isolates, 35.7%), S. pneumoniae (128, 12.9%), M. catarrhalis (114, 11.5%), E. coli (89, 8.9%), S. aureus (89, 8.9%), K. pneumoniae (82, 8.2%), A. baumannii (31, 3.1%), P. aeruginosa (28, 2.8%), Enterobacter cloacae (18, 1.8%), S. agalactiae (13, 1.3%), Enterobacter aerogenes (12, 1.2%), Klebsiella oxytoca (9, 0.9%), Serratia marcescens (6, 0.6%), Citrobacter koseri (5, 0.5%), Acinetobacter junii (3, 0.3%), S. haemolyticus (3, 0.3%), Pseudomonas luteola (2, 0.2%), Raoultella planticola (2, 0.2%), and one each of Enterococcus faecalis, Serratia fonticola, Pseudomonas mendocina, Proteus mirabilis, Enterobacter gergoviae and Salmonella spp, each accounting for 0.1%, respectively (Table S1).
Antimicrobial Susceptibility Profile
The commonly prescribed antimicrobial drugs for clinical use in China were selected to determine the susceptibility pattern for each bacterial isolates obtained in this study. A total of 702 strains (70.5%) showed with the resistant (R) and/or intermediate (I) phenotypes to at least one of the tested drugs. The details are follows.
Gram-Positive Bacteria
All strains of the five Gram-positive bacteria (S. pneumoniae, S. aureus, S. agalactiae, S. haemolyticus and E. faecalis) were tested for their susceptibilities to antibiotics by the VITEK 2 Compact automatic system with GP cards. The summary results of antimicrobial susceptibility testing of these five species are presented in Table 1. Below are brief descriptions for each of the five species.
S. pneumoniae
All the 128 S. pneumoniae strains were tested for their susceptibilities to the following seven antibiotics: penicillin, erythromycin, trimethoprim-sulfamethoxazole, vancomycin, linezolid, meropenem, and ceftriaxone (Table 1). All the 128 S. pneumoniae strains were resistant to erythromycin. The rates of resistance to trimethoprim-sulfamethoxazole and ceftriaxone were 93.8% (120/128), and 39.8% (51/128), respectively. All the S. pneumoniae strains were sensitive to penicillin, vancomycin, linezolid, and meropenem (Table 1).
S. aureus and S. haemolyticus
All the 89 S. aureus strains and three S. haemolyticus strains were tested for their susceptibilities to the following seven antibiotics: penicillin, oxacillin, erythromycin, clindamycin, trimethoprim-sulfamethoxazole, vancomycin, and linezolid. The S. aureus and S. haemolyticus strains showed different resistance rates to five of the seven drugs but all strains were sensitive to linezolid, and vancomycin. The AMR rates of S. aureus to the five drugs, from the highest to lowest, were penicillin (78.7%, 70/89), erythromycin (24.7%, 22/89), oxacillin (MRSA) (22.5%, 20/89), clindamycin (22.5%, 20/89), and trimethoprim-sulfamethoxazole (7.9%, 7/89) (Table 2). All three S. haemolyticus strains were resistant to penicillin, erythromycin, and oxacillin, two S. haemolyticus strains were resistant to trimethoprim-sulfamethoxazole and one S. haemolyticus strain was resistant to clindamycin (Table 1).
S. agalactiae and E. faecalis
All the 13 S. agalactiae strains were tested for their susceptibilities to the following five antibiotics: penicillin, ampicillin, clindamycin, vancomycin, and linezolid. Our S. agalactiae population showed a high resistance rate to clindamycin (92.3%, 12/13), but all 13 strains were sensitive to the other four drugs (Table 1). The E. faecalis strain was sensitive to all the tested five drugs: penicillin, ampicillin, erythromycin, vancomycin, and gentamicin (Table 1).
Gram-Negative Bacteria
The H. influenzae and M. catarrhalis strains were all tested for their susceptibility to antibiotics by the KB disk diffusion method. For other Gram-negative bacterial strains, including those of E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, E. cloacae, E. aerogenes, K. oxytoca, S. marcescens, C. koseri, A. junii, P. Luteola, R. planticola, S. fonticola, P. mendocina, P. mirabilis, E. gergoviae and Salmonella spp. their antibiotic susceptibilities were determined by the VITEK 2 Compact automatic system with GN cards. The summary results of these tests are presented in Figures 4, 5, and Table 2.
H. influenzae and M. catarrhalis
All the 356 H. influenzae strains were tested for their susceptibilities to the following seven antibiotics: ampicillin, amoxicillin-clavulanic acid, ceftriaxone, cefuroxime, meropenem, trimethoprim-sulfamethoxazole, and azithromycin (Figure 4). Among the 356 strains, 322 (90.4%) showed the R and/or I phenotypes to at least one of the seven drugs. The overall resistance rates of H. influenzae to the seven drugs, from the highest to lowest, were trimethoprim-sulfamethoxazole (80.3%, 286/356), ampicillin (76.7%, 273/356), cefuroxime (58.4%, 208/356), azithromycin (29.5%, 105/356), amoxicillin-clavulanic acid (18.3%, 65/356), ceftriaxone (5.3%, 19/356), and meropenem (0.6%, 2/356) (Figure 4).
All the 114 M. catarrhalis strains were tested for their susceptibilities to the following six drugs: amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole, azithromycin, erythromycin, clarithromycin, and tetracycline (Figure 5). All the M. catarrhalis strains were sensitive to tetracycline and clarithromycin, while the M. catarrhalis strains showed different resistance rates to other four drugs: amoxicillin-clavulanic acid (1.8%, 2/114), trimethoprim-sulfamethoxazole (29.8%, 34/114), azithromycin (41.2%, 47/114), and erythromycin (44.7%, 51/114) (Figure 5). In addition, 60.5% (69/114) of M. catarrhalis strains and 42.9% (153/356) of H. influenzae strains produced β-lactamases.
E. coli
All the 89 E. coli strains were tested for their susceptibilities to the following 12 antibiotics: ampicillin, ampicillin-sulbactam, piperacillin-tazobactam, cefazolin, ceftriaxone, ceftazidime, cefepime, imipenem, aztreonam, trimethoprim-sulfamethoxazole, tobramycin, and imipenem (Table 2). The E. coli strains showed different resistance rates to 10 of the 12 antibiotics except for amikacin and piperacillin-tazobactam where no resistant strain was isolated. The resistance rates of E. coli to the 10 drugs from the highest to lowest were ampicillin (68.5%, 61/89), ampicillin-sulbactam (52.8%, 47/89), trimethoprim-sulfamethoxazole (29.2%, 26/89), cefazolin (23.6%, 21/89), ceftriaxone (21.4%, 19/89), tobramycin (21.4%, 19/89), ceftazidime (20.2%, 18/89), aztreonam (19.1%, 17/89), cefepime (1.1%, 1/89), and amikacin (1.1%, 1/89). In addition, 19.1% (17/89) E. coli strains produced extended spectrum β-lactamases.
K. pneumoniae and K. oxytoca
All the 82 K. pneumoniae strains and nine K. oxytoca strains were tested for their susceptibilities to the following 11 antibiotics: ampicillin-sulbactam, piperacillin-tazobactam, cefazolin, ceftriaxone, ceftazidime, cefepime, imipenem, aztreonam, trimethoprim-sulfamethoxazole, tobramycin, and amikacin (Table 2). All the K. pneumoniae strains were sensitive to amikacin. One each of the 82 K. pneumoniae strains was resistant to piperacillin-tazobactam, cefepime, imipenem (CRE) and piperacillin-tazobactam respectively. The resistance rates of K. pneumoniae to the other six drugs from the highest to lowest were ampicillin-sulbactam (18.3%, 15/82), cefazolin (13.4%, 11/82), trimethoprim-sulfamethoxazole (13.4%, 11/82), ceftazidime (10.9%, 9/82), ceftriaxone (8.5%, 7/82), and aztreonam (4.9%, 4/82). The CRE strains were resistant to nine of the 11 drugs except tobramycin and amikacin. 6.1% (5/82) K. pneumoniae produced extended-spectrum β-lactamases. Three of the nine (33.3%) K. oxytoca strains were resistant to cefazolin. All the nine K. oxytoca strains were sensitive to the other ten drugs.
A. baumannii and A. junii
All the 31 A. baumannii strains and three A. junii were tested for their susceptibilities to the following eight antibiotics: ampicillin-sulbactam, ceftriaxone, ceftazidime, cefepime, imipenem, trimethoprim-sulfamethoxazole, tobramycin, and amikacin (Table 2). Two of the 31 (6.5%) A. baumannii strains were resistant to trimethoprim-sulfamethoxazole, 11 (35.5%) were resistance to ceftriaxone, while the other A. baumannii strains were sensitive to the other six drugs. All three A. junii strains were sensitive to all the eight drugs.
P. aeruginosa, P. luteola and P. mendocina
All the 28 P. aeruginosa strains were tested for their susceptibilities to the following six antibiotics: piperacillin-tazobactam, ceftazidime, cefepime, imipenem, tobramycin, and amikacin (Table 2). Two P. luteola and one P. mendocina strains were tested for their susceptibilities to the following eight antibiotics: piperacillin-tazobactam, ceftriaxone, ceftazidime, cefepime, imipenem, trimethoprim-sulfamethoxazole, tobramycin, and amikacin. The P. mendocina strain was resistant to ceftriaxone and ceftazidime. The other tested strains were all sensitive to the tested drugs.
E. cloacae, E. aerogenes and E. gergoviae
All the 18 E. cloacae, 12 E. aerogenes and one E. gergoviae strains were tested for their susceptibilities to the following nine antibiotics: piperacillin-tazobactam, ceftriaxone, ceftazidime, cefepime, imipenem, aztreonam, trimethoprim-sulfamethoxazole, tobramycin, and amikacin (Table 2). One E. aerogenes isolate was resistant to ceftriaxone, ceftazidime, cefepime, and aztreonam. Other tested strains were all sensitive strains to all tested antibiotics.
S. marcescens and S. fonticola
All the six S. marcescens and one S. fonticola strains were tested for their susceptibilities to the same antibiotics as Enterobacter sp. above (Table 2). The S. fonticola strain was resistant to ceftriaxone, ceftazidime, and aztreonam but sensitive to other six drugs. All the six S. marcescens strains were sensitive to the nine tested antibiotics.
Others
Five C. koseri, two R. planticola, one P. mirabilis, and one Salmonella strains were tested for their susceptibilities to different drugs (Table 2). All the strains were sensitive to the tested drugs except the P. mirabilis strain which was resistant to trimethoprim-sulfamethoxazole.
Discussion
Pneumonia is a major cause of morbidity and mortality. According to the 2015 and 2016 GBD data, the main causes of pneumonia deaths in children were bacteria.1,27 Similar to those reported from Chengdu (southwest China) and Xiamen (southeast China),25,28 the detected bacterial species in this study mainly belonged to Gram-negative taxa, and that the 1 month to 1 year old and 2 to 5 years old groups had higher bacteria carriage rates than those of less than 1 month and the 6 to 14 years old groups (p = 0.000). However, no statistically significant difference in bacterial species composition and isolate rate was found between the two sex groups. Our results are different from a study in India that demonstrated boys under 5 years old had a higher incidence of respiratory tract infection than girls.29
H. influenzae, S. pneumoniae, and M. catarrhalis are the top three bacterial pathogens colonizing the lower respiratory tract of children with pneumonia. A previous study showed that these three bacteria were positively correlated with each other in their carriage rates at the population level.30 However, differences among age groups were found for several species, similar to two previous reports.27,31 For example, in our sample, H. influenzae (55.2%) were mainly isolated from the 2 to 5 years old group while only 3.6% was found in the less than 1 month age group. In contrast, samples from less than 1 month old had a significantly higher percentage of E. coli (27.0%) than those from 2 to 5 years old (0.3%) and 6 to 14 years old (0.0%).
Our results also observed significant influence of seasons on species distribution. For example, the samples from spring and winter had higher percentages of H. influenzae (45.0% and 47.4%) than those from summer and autumn (25.6% and 24.4%). Our results contrast those reported recently of a retrospective study from Chengdu, China. In that study, H. influenzae was detected more frequently in spring than other species, while S. pneumoniae and M. catarrhalis were detected more frequently in winter.27 Seasonal differences have been found in many fungal pathogens, largely related to differences in temperature and moisture among seasons and the impact of those differences on fungal growth and reproduction in both our indoor and outdoor living environments.10,32
AMR is a recognized global threat and increases the risk of infection-related death for patients of all age groups, including children. In our study, a total of 702 strains (70.5%) showed the R and/or I phenotypes to at least one of the tested drugs in this study. Our results suggest that susceptibility testing should be implemented before antibiotic treatment in Hainan to improve treatment success. The main bacteria detected in our cohort was H. influenzae and we observed high AMR rates of this species to sulfamethoxazole (80.3%) and ampicillin (76.9%), similar to those reported in southern parts of Mainland China27,28 Ampicillin is a historical drug of choice for treating H. influenzae infection, but in recent years, ampicillin resistance has increased substantially,33–37 suggesting that ampicillin should no longer be the first-line treatment for infections caused by H influenzae in China. Furthermore, strains of H. influenzae isolated here showed variable rates of resistance to 5 other commonly used antibiotics (Figure 4), with 42.9% of these strains producing β-lactamases. Interestingly, though the rate of β-lactamase production was high, it was lower than that reported in a national survey in China by the Infectious Disease Surveillance of Pediatrics (ISPED) program during 2016–2020.38
The second most common bacterium was S. pneumoniae and we observed all strains of this species being resistant to erythromycin. In addition, we observed a high frequency resistance to trimethoprim-sulfamethoxazole. Interestingly, there was a low frequency of resistance to ceftriaxone and all S. pneumoniae strains in our collection were extremely sensitive to penicillin, vancomycin, linezolid, and meropenem (Table 2). These data are generally consistent with those reported previously.27,28,35 Together, our results suggest that penicillin, vancomycin, linezolid, and meropenem can be used as the first-line antibiotics for the treatment of pneumoniae caused by S. pneumoniae in Hainan.
The third most common bacterial species was M. catarrhalis and we observed that 60.5% of M. catarrhalis strains produced β-lactamases. This rate is far lower than previously reported by ISPED (>95%).37 However, all the M. catarrhalis strains were sensitive to tetracycline and clarithromycin, and with different degrees of resistance to amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole, azithromycin, and erythromycin.
Among the 89 S. aureus and three S. haemolyticus strains isolated in our study, 22.5% and 100% were resistant to oxacillin, respectively. A similar rate was reported by Zhu et al during 2018–2020 where 27% of S. aureus detected in the lower respiratory tract of children in Chengdu, southwest China were MRSA and with an increasing trend of resistance over the three years.27 Reports from Japan suggested that the rapid spread of S. aureus carrying the resistance gene in the environment was associated with increased MRSA detection rate in hospitals.39 Similarly, reports from Spain showed that the use of antibiotics in animal husbandry promoted the spread of Staphylococcus antibiotic resistance genes in the environment and wildlife.40 We also found the oxacillin-resistant S. aureus and oxacillin-resistant S. haemolyticus strains were also resistant to penicillin and were more likely to be resistant to other antibiotics than the oxacillin-sensitive S. aureus and oxacillin-sensitive S. haemolyticus. Interestingly, all S. aureus and S. haemolyticus strains in our study were sensitive to vancomycin and linezolid. Our results suggest that these two drugs can be used as first-line drugs for treating infections caused by MRSA and MRSH in Hainan.
E. coli and K. pneumoniae were the most common species producing extended-spectrum β-lactamases. In this study, we found that the frequency of extended-spectrum β-lactamases production from E. coli and K. pneumoniae were 19.1% and 6.1%, respectively. These rates are far lower than those reported in the previous studies.41,42 Studies conducted by Wang et al43 and Rodriguez-Bano et al44 showed statistically significant relationship between age groups and the percentage of extended-spectrum β-lactamases producers, with the oldest age group having a greater proportion of extended-spectrum β-lactamases-producing strains. Over half of the E coli strains in our sample were resistant to ampicillin and ampicillin-sulbactam, similar to those reported in a previous study.28 However, our E. coli sample showed a lower frequency of resistance to other 10 antibiotics (trimethoprim-sulfamethoxazole, cefazolin, ceftriaxone, tobramycin, ceftazidime, aztreonam, cefepime, amikacin, and piperacillin-tazobactam) than previous reports27,28,38 (Table 2). Among all strains isolated here, the CRE strain showed the broadest antibiotic resistance profile, being resistant to nine of the 11 tested drugs except tobramycin and amikacin (Table 2).
Conclusion
In conclusion, our analyses of the distribution and drug resistance characteristics of bacterial pathogens revealed significant age and seasonal patterns of bacterial infection epidemiology in children with pneumonia in Hainan. High drug resistance rates were observed for most bacterial species to several of the antibiotics. Our results call for greater effects in vaccination, hand hygiene, strengthening of personal protective measures, and aseptic operation of invasive medical treatment to reduce the spread of drug-resistant pathogens. In addition, the highly variable AMR rates among the pathogen species-antibiotic combinations call for the development of rapid and accurate detection of both the pathogens and their antibiotic susceptibilities in individual jurisdictions.37 Such data could help shorten decision-making and improve treatment outcomes against these pathogens.
Abbreviations
MDR, Multidrug-resistant; MRSA, methicillin (oxacillin) resistant Staphylococcus aureus; MRSH, methicillin (oxacillin) resistant Staphylococcus haemolyticus; CRE, Carbapenem resistant Enterobacteriaceae; VRE, Vancomycin resistant Enterococci; KB, Kirby-Bauer disk diffusion; CLSI, Clinical and Laboratory Standards Institute; AMR, Antimicrobial resistance; ISPED, Infectious Disease Surveillance of Pediatrics; H. influenza, Hemophilus influenza; S. pneumoniae, Streptococcus pneumoniae; M. catarrhalis, Moraxella catarrhalis; E. coli, Escherichia coli; S. aureus, Staphylococcus aureus; K. pneumoniae, Klebsiella pneumoniae; A. baumannii, Acinetobacter baumannii; P. aeruginosa, Pseudomonas aeruginosa; P. aeruginosa, Pseudomonas aeruginosa; S. agalactiae, Streptococcus agalactiae; S. agalactiae, Streptococcus agalactiae; E. aerogenes, Enterobacter aerogenes; K. oxytoca, Klebsiella oxytoca; S. marcescens, Serratia marcescens; C. koseri, Citrobacter koseri; A. junii, Acinetobacter junii; P. luteola, Pseudomonas luteola; R. planticola, Raoultella planticola; E. faecalis, Enterococcus faecalis; S. fonticola, Serratia fonticola; P. mendocina, Pseudomonas mendocina; P. mirabilis, Proteus mirabilis; E. gergoviae, Enterobacter gergoviae; E. cloacae, Enterobacter cloacae.
Ethics Statement
The study complied with the Declaration of Helsinki.
Funding
Financial support for this project came from the National Natural Science Foundation of China (Grant No. 31860035), and Natural Science Foundation of Hainan Province (Grant No.822RC708, 2019RC227 and 819MS142).
Disclosure
The authors report no conflicts of interest in this work.
References
1. McAllister DA, Liu L, Shi T, et al. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob Health. 2019;7(1):e47–e57. doi:10.1016/S2214-109X(18)30408-X
2. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the Sustainable Development Goals [published correction appears in Lancet. 2017 May 13;389(10082):1884]. Lancet. 2016;388(10063):3027–3035. doi:10.1016/S0140-6736(16)31593-8
3. Feng XL, Theodoratou E, Liu L, et al. Social, economic, political and health system and program determinants of child mortality reduction in China between 1990 and 2006: a systematic analysis. J Glob Health. 2012;2:010405. doi:10.7189/jogh.02.010405
4. World Health Organization. Global and Regional Immunization Profile. Geneva: World Health Organization; 2017.
5. Rudan I, O’Brien KL, Nair H, et al. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health. 2013;3(1):010401. doi:10.7189/jogh.03.010401
6. Zar HJ, Andronikou S, Nicol MP. Advances in the diagnosis of pneumonia in children. BMJ. 2017;358:j2739. doi:10.1136/bmj.j2739
7. Chintu C, Mudenda V, Lucas S, et al. Lung diseases at necropsy in African children dying from respiratory illnesses: a descriptive necropsy study. Lancet. 2002;360(9338):985–990. doi:10.1016/S0140-6736(02)11082-8
8. Santoso P, Sung M, Hartantri Y, et al. MDR pathogens organisms as risk factor of mortality in secondary pulmonary bacterial infections among COVID-19 patients: observational studies in two referral hospitals in West Java, Indonesia. Int J Gen Med. 2022;15:4741–4751. doi:10.2147/IJGM.S359959
9. Huemer M, Mairpady Shambat S, Brugger SD, Zinkernagel AS. Antibiotic resistance and persistence-implications for human health and treatment perspectives. EMBO Rep. 2020;21(12):e51034. doi:10.15252/embr.202051034
10. Xu J. Assessing global fungal threats to humans. mLife. 2022;1(3):223–240. doi:10.1002/mlf2.12036
11. Woolhouse M, Waugh C, Perry MR, Nair H. Global disease burden due to antibiotic resistance - state of the evidence. J Glob Health. 2016;6:010306. doi:10.7189/jogh.06.010306
12. Tiri B, Sensi E, Marsiliani V, et al. Antimicrobial stewardship program, COVID-19, and infection control: spread of carbapenem-resistant Klebsiella pneumoniae colonization in ICU COVID-19 patients. What did not work? J Clin Med. 2020;9:E2744.
13. Li J, Wang J, Yang Y, et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9:153. doi:10.1186/s13756-020-00819-1
14. Contou D, Claudinon A, Pajot O, et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10:119. doi:10.1186/s13613-020-00736-x
15. Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:646. doi:10.1186/s12879-020-05374-z
16. Fu Y, Yang Q, Xu M, et al. Secondary bacterial infections in critical ill patients with coronavirus disease 2019. Open Forum Infect Dis. 2020;7:ofaa220.
17. Nori P, Szymczak W, Puius Y, et al. Emerging co-pathogens: New Delhi metallo-β-lactamase producing Enterobacteriaceae infections in New York City COVID-19 patients. Int J Antimicrob Agents. 2020;106179. doi:10.1016/j.ijantimicag.2020.106179
18. Farfour E, Lecuru M, Dortet L, et al. Carbapenemase-producing Enterobacterales outbreak: another dark side of COVID-19. Am J Infect Control. 2020;48:1533–1536. doi:10.1016/j.ajic.2020.09.015
19. Posteraro B, Torelli R, Vella A, et al. Pan-echinocandin-resistant Candida glabrata bloodstream infection complicating COVID-19: a fatal case report. J Fungi. 2020;6:163. doi:10.3390/jof6030163
20. Chowdhary A, Tarai B, Singh A, Sharma A. Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April–July 2020. Emerg Infect Dis. 2020;26:2694–2696. doi:10.3201/eid2611.203504
21. Mohamed A, Hassan T, Trzos-Grzybowska M, et al. Multi-triazole-resistant Aspergillus fumigatus and SARS-CoV-2 co-infection: a lethal combination. Med Mycol Case Rep. 2021;31:11–14. doi:10.1016/j.mmcr.2020.06.005
22. Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26:1395–1399. doi:10.1016/j.cmi.2020.06.025
23. Lai CC, Chen SY, Ko WC, Hsueh PR. Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents. 2021;57(4):106324. doi:10.1016/j.ijantimicag.2021.106324
24. Centers for Disease Control and Prevention. Multidrug-resistant organisms (MDRO) management. Available from: https://www.cdc.gov/infectioncontrol/guidelines/mdro/.
25. Zhu X, Ye T, Zhong H, et al. Distribution and drug resistance of bacterial pathogens associated with lower respiratory tract infection in children and the effect of COVID-19 on the distribution of pathogens. Can J Infect Dis Med Microbiol. 2022;1181283. doi:10.1155/2022/1181283
26. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
27. GBD 2016 Lower Respiratory Infections Collaborators Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. doi:10.1016/S1473-3099(18)30310-4
28. Su DQ, Huang HL, Zhuo ZQ. Pathogen distribution and bacterial resistance in children with severe pneumonia: a single-center retrospective study. Medicine. 2021;100(35):e27128. doi:10.1097/MD.0000000000027128
29. Krishnan A, Amarchand R, Gupta V, et al. Epidemiology of acute respiratory infections in children - preliminary results of a cohort in a rural north Indian community. BMC Infect Dis. 2015;15:462. doi:10.1186/s12879-015-1188-1
30. Dunne EM, Murad C, Sudigdoadi S, et al. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: a cross-sectional study. PLoS One. 2018;13(4):e0195098. doi:10.1371/journal.pone.0195098
31. Musher DM, Jesudasen SS, Barwatt JW, Cohen DN, Moss BJ, Rodriguez-Barradas MC. Normal respiratory flora as a cause of community-acquired pneumonia. Open Forum Infect Dis. 2020;7(9):ofaa307. doi:10.1093/ofid/ofaa307
32. Xu J. Origins and spread of plant fungal and oomycete disease outbreaks. J Plant Protect. 2022;49(1):283–297.
33. Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis. 2011;53(7):e25–e76. doi:10.1093/cid/cir531
34. Jochem WC, Razzaque A, Root ED. Effects of health intervention programs and arsenic exposure on child mortality from acute lower respiratory infections in rural Bangladesh. Int J Health Geogr. 2016;15(1):32. doi:10.1186/s12942-016-0061-9
35. Troeger C, Forouzanfar M, Rao PC; GBD 2015 LRI Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17(11):1133–1161. doi:10.1016/S1473-3099(17)30396-1
36. Torumkuney D, Chaiwarith R, Reechaipichitkul W, et al. Results from the Survey of Antibiotic Resistance (SOAR) 2012–14 in Thailand, India, South Korea and Singapore. J Antimicrob Chemother. 2016;71(Suppl1):i3–i19. doi:10.1093/jac/dkw073
37. Vaez H, Sahebkar A, Pourfarzi F, Yousefi-Avarvand A, Khademi F. Prevalence of antibiotic resistance of Haemophilus influenzae in Iran- a meta-analysis. Iran J Otorhinolaryngol. 2019;31(107):349–357. doi:10.22038/ijorl.2019.34363.2137
38. Fu P, Xu H, Jing C, et al. Bacterial epidemiology and antimicrobial resistance profiles in children reported by the ISPED program in China, 2016 to 2020. Microbiol Spectr. 2021;9(3):e0028321. doi:10.1128/Spectrum.00283-21
39. Mitsumoto-Kaseida F, Murata M, Toyoda K, et al. Clinical and pathogenic features of SCCmec type II and IV methicillin-resistant Staphylococcus aureus in Japan. J Infect Chemother. 2017;23(2):90–95. doi:10.1016/j.jiac.2016.11.001
40. García LA, Torres C, López AR, Rodríguez CO, Espinosa JO, Valencia CS. Staphylococcus spp. from wild mammals in Aragón (Spain): antibiotic resistance status. J Vet Res. 2020;64(3):373–379. doi:10.2478/jvetres-2020-0057
41. Hu F, Wang F, Jiang X, et al. Report of CHINET antimicrobial resistance surveillance program in 2015. Chin J Infect Chemother. 2016;16(6):685–694.
42. Hu F, Zhu D, Wang F, et al. CHINET surveillance of bacterial resistance across China: report of the results in 2016. Chin J Infect Chemother. 2016;17(5):481–491.
43. Wang Y, Zhang Q, Jin Y, Jin X, Yu J, Wang K. Epidemiology and antimicrobial susceptibility profiles of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli in China. Braz J Microbiol. 2019;50(3):669–675. doi:10.1007/s42770-019-00081-7
44. Rodríguez-Baño J, Alcalá JC, Cisneros JM, et al. Community Infections Caused by Extended-Spectrum β-Lactamase–Producing Escherichia coli. Arch Intern Med. 2008;168(17):1897–1902. doi:10.1001/archinte.168.17.1897
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


