Back to Journals » International Journal of General Medicine » Volume 16
Autoimmune Disease-Related Hub Genes are Potential Biomarkers and Associated with Immune Microenvironment in Endometriosis
Authors Yang YT , Jiang XY , Xu HL , Chen G , Wang SL, Zhang HP , Hong L, Jin QQ , Yao H, Zhang WY , Zhu YT, Mei J, Tian L , Ying J , Hu JJ, Zhou SG
Received 21 April 2023
Accepted for publication 30 June 2023
Published 10 July 2023 Volume 2023:16 Pages 2897—2921
DOI https://doi.org/10.2147/IJGM.S417430
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Yin-Ting Yang,1,2,* Xi-Ya Jiang,1,2,* Hong-Liang Xu,3,* Guo Chen,1,2,* Sen-Lin Wang,4 He-Ping Zhang,3 Lin Hong,1,2 Qin-Qin Jin,1,2 Hui Yao,1,2 Wei-Yu Zhang,1,2 Yu-Ting Zhu,1,2 Jie Mei,1,2 Lu Tian,1,2 Jie Ying,1,2 Jing-Jing Hu,5 Shu-Guang Zhou1,2
1Department of Gynecology and Obstetrics, Maternity and Child Healthcare Hospital Affiliated to Anhui Medical University, Anhui Province Maternity and Child Healthcare Hospital, Hefei, Anhui, 230001, People’s Republic of China; 2Department of Gynecology and Obstetrics, The Fifth Clinical College of Anhui Medical University, Hefei, Anhui, 230032, People’s Republic of China; 3Department of Pathology, Anhui Province Maternity and Child Healthcare Hospital, Hefei, Anhui, 230001, People’s Republic of China; 4Department of Medical Genetics Center, Anhui Province Maternity and Child Healthcare Hospital, Hefei, Anhui, 230001, People’s Republic of China; 5Department of Gynecology and Obstetrics, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shu-Guang Zhou, Department of Gynecology, Anhui Medical University Affiliated Maternity and Child Healthcare Hospital, The Fifth Clinical College of Anhui Medical University, Anhui Province Maternity and Child Healthcare Hospital, No. 15, Yi-Min Street, Lu-Yang District, Hefei, Anhui, 230001, People’s Republic of China, Email [email protected] Jing-Jing Hu, Department of Gynecology and Obstetrics, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China, Email [email protected]
Background: Endometriosis, a common gynecological condition, can cause symptoms such as dysmenorrhea, infertility, and abnormal bleeding, which can negatively affect a woman’s quality of life. In the current study, the pathophysiological mechanisms of endometriosis are unknown, but this study suggests that endometriosis is associated with dysregulation of the autoimmune system. This study identify hub genes involved in the prevalence, identification and diagnostic value of endometriosis and autoimmune diseases, and explore the central genes and immune infiltrates, the diagnosis of endometriosis provides a new sight of thinking about diagnosis and treatment.
Methods and Results: The relevant datasets for endometriosis GSE141549, GSE7305 and autoimmune disease-related genes (AIDGs) were downloaded from online database. Using the “limma” package and WGCNA to screen out the autoimmune disease related genes and endometriosis related genes, the autoimmune disease gene-related differential genes (AID-DEGs) progressive GO, KEGG enrichment analysis, and then using the protein interaction network and Cytoscape software to select hub genes (CXCL12, PECAM1, NGF, CTGF, WNT5A), using the “pROC” package to analyze the hub genes for the diagnostic value of endometriosis. The difference in the importance of hub genes for the diagnosis of endometriosis was analyzed by machine learning random forest, and the combined diagnostic value of hub genes was analyzed by using the Support Vector Machine (SVM) algorithm. The eutopic (EU) and ectopic endometrium (EC) immune microenvironment of endometriosis was evaluated using CIBERSORT, the correlation of hub genes to the immune microenvironment was analyzed.
Conclusion: The hub genes associated with AIDGs are differentially expressed in EC and EU of endometriosis and possess important value for the diagnosis of endometriosis. The hub genes have a very important impact on the immune microenvironment of endometriosis, which is important for exploring the connection between endometriosis and autoimmune diseases and provides a new insight for the subsequent study of immunotherapy and diagnosis of endometriosis.
Keywords: EMT, hub genes, diagnosis, immunotherapy, immune microenvironment
Introduction
Endometriosis (EMT) is a disease in which endometrium-like tissue grows outside the endometrium. EMT is an inflammatory endometrial disease that affects women of reproductive age and is progesterone and oestrogen dependent.1 Approximately 200 million women and teens globally are affected by it, or between 7 and 10% of women of reproductive age.2,3 The staging of endometriosis is based on the location, number, size and degree of adhesions of the ectopic endometrium, but the severity of symptoms or recurrence of endometriosis is not related to stage. For example, women with revised American Society of Reproductive Medicine (rASRM) stage I disease may have severe pain, infertility, or both, whereas stage IV endometriosis may be asymptomatic. When symptoms of EMT are severe, it can cause recurrent dysmenorrhoea, persistent pelvic pain, abnormal menstruation and non-uterine amenorrhoea, leading to infertility in women.4 According to some reports, EMT affects 71% to 87% of women who experience chronic pelvic pain.2
The current ideas for understanding the origins of EMT are as following, Sampson’s “menstrual blood reverse flow” theory was initially put forth in 1927. Sampson came to believe that the majority of cases of EMT were brought on by endometrial fragments that were absorbed through the fallopian duct during menstruation, either by change from endometrial to ectopic endometrium, and by peritoneal and ovarian growth.5 The word “incumbent intimal determinism”, which refers to the patient’s incumbent endometrial traits that influence whether the endometrial fragments can develop in ectopic plants by blood reflux, was first proposed by Lang et al.6 It’s possible that there’s a correlation between EMT and the body’s immune mechanism dysfunction, according to pathophysiology ideas like autoantibodies7 and autoimmune regulation.8 Normal human immune system modulation can quickly identify and eliminate foreign objects and lessen inflammatory reactions. Therefore, it is believed that immune disorder is the root cause of EMT. Other studies have shown that genetic mutations are associated with the development of EMT, such as the dramatic increase in mutant allele frequencies (MAF) for Kirsten rats arcomaviral oncogene homolog (KRAS) mutations in endometriotic epithelium suggests that endometrial tissue carrying KRAS mutations is transported in a retrograde direction to the ovarian surface, where specific KRAS mutations give them a selective advantage at that tissues and other ectopic tissues, leading to the development and widespread distribution of EMT.9 Other theories of pathogenesis are the stem cell theory;10 malignant tendency;11 and so forth.
Studies illustrated that there are correlations between autoimmune diseases (AID) and the occurrence of EMT.12 A number of studies have found a significantly higher prevalence of autoimmune thyroid disease in women with EMT than in controls.13 One of the potential associations is autoimmunity. EMT shares many features with autoimmune diseases, including polyclonal B-cell activation, abnormal T- and B-cell function, and inflammatory tissue damage.14 Humoral immunity also explains its association with autoimmune thyroid disease. A high reactivity of some autoantibodies, such as thyroid peroxidase antibodies (TPOAb), has been found in patients with EMT. In addition, the incidence of positive antinuclear antibodies (ANAs) appears to be higher in patients with EMT than in patients with Graves’ disease. Another possible link between Graves’ disease and EMT is the estrogenic effect:15 Graves’ disease is five times more common in women than in men. Estrogen is an important regulator of the immune system, acting as a regulator of cytokine expression, antigen presentation and B-cell lymphocyte production. In addition, a large number of clinical studies have confirmed that the incidence of autoimmune thyroid diseases, systemic lupus erythematosus,16 inflammatory bowel disease and rheumatoid arthritis in EMT patients is significantly higher than that of normal people,17 and there are correlations in pathogenesis, clinical symptoms, disease progression and prognosis.18
This study used Weighted gene co-expression network analysis (WGCNA) for feature selection, screening for autoimmune disease related genes (AIDGs) involved in EMT, and verifying the diagnostic value of hub genes in EMT through machine learning algorithms. At the same time, this study also revealed the specific impact of AIDGs hub genes on the changes in the immune microenvironment of EMT patients.
Methods and Materials
Gene Expression Data and Autoimmune Disease Related Gene
A public functional genomics database called Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) offers high-throughput data for gene expression and microarray analysis. The EMT-related datasets (GSE141549 and GSE7305) were downloaded from GEO database.19,20 All datasets are belonged to Homo sapiens; the GSE141549 data platform was GPL13376, and the GSE7305 data platform was GPL570. GSE141549 contained 338 samples, 61 eutopic (EU) and 175 ectopic (EC) endometrium samples are retained by screening the sample, and GSE7305 contained 20 samples, including 10 EC and 10 EU samples. The current investigation comprised all samples from these two datasets; GSE141549 was used to screen for the hub gene, and GSE7305 was utilized to confirm the diagnostic value. Standardized data processing was carried out using the R package “limma”.21 A total of 4186 AIDGs were downloaded from the AIDGs Database (GAAD, http://gaad.medgenius.info/genes/).
Identification of DEGs
The samples were divided into an EC group and an EU group based on the expression matrix generated from the dataset GSE141549. Using the R package “limma” with absolute values of log fold-change (log FC) > 1 and adj.P.Val 0.05 as thresholds, differentially expressed genes (DEGs) between the several groups were found. In the EC group, DEGs that had elevated and downregulated logFC values, respectively, were found. The DEG analysis results are presented as volcano maps created with the “ggplot2” R package.22
Weighted Gene Co-Expression Network Analysis
The GSE141549 dataset’s expression data were analyzed using WGCNA R package, and a co-expression network using EC and EU samples as clinical characteristics was created. In order to identify and eliminate outliers, sample clustering was carried out using the “goodSamplesGenes” function. The WGCNA package’s “pickSoftThreshold” function was used to determine the soft threshold’s optimal filtering power value, and the “blockwiseModules” function was used to create the one-step co-expression matrix. The adjacency matrix was obtained by first calculating the adjacency of the expression levels across all genes. Next, the adjacency matrix can be converted into a topological overlap matrix (topological overlap matrix, TOM), and calculate the corresponding difference degree (1-TOM), and divide the module according to the dynamic shear tree standard. When 1-TOM is used as the measurement value, the minimum capacity of the module is set to 30 and the shear height of the module is 0.20. With a minimum of 100 genes in each module, the dynamic tree cutting method was employed to identify various modules. Then, to merge related modules, a threshold of 0.2 was chosen. The implementation of a correlation analysis between module membership and gene significance followed. The key module genes, differential genes (DEGs) and AIDGs set were screened and crossed to obtain the autoimmune disease gene-related differential genes (AID-DEGs). In addition, Venn diagrams were used to screen to hub genes.
Functional Enrichment Analysis
The AID-DEGs were assessed using the GO functional enrichment research, which includes biological process (BP), molecular function (MF), cellular component (CC), and KEGG pathway analysis. KEGG is a database that provides information on the molecular and higher-level functions of genes, including biochemical pathways. The “clusterProfiler” R package handled the annotation and visualization.23 A hypergeometric test was used to do the enrichment analysis. The cut-off number for a statistically significant difference was set at P<0.05. “GOplot” packages were used to screen for enrichment of genes with more than 40 replicates in the enrichment pathway in the first 6 biological pathways.
Construction of the Protein–Protein Interaction Network
The STRING database (https://string-db.org) was utilized to generate the protein-protein interaction network (PPI) of the altered genes.24 “Medium confidence (0.400)” interaction scores were established as the minimum requirements. The PPI network was then visualized using Cytoscape software.25 Important PPI network modules were also found using the molecular complex detection (MCODE)26 plug-in with degree cutoff = 2, maximum depth = 100, and k-score = 2. The Hub Genes were discovered via Cytohubba,27 a plug-in for the Cytoscape software.
Analyze the Differences in Expression of Hub Genes and Their Diagnostic Value
Analyze the difference of hub gene expression in EC group and EU group by the “ggpubr” package, and “pROC” package28 was used to analyze the diagnostic value of hub gene in EC group and EU group. GSE7305 as the verification set for verification the differences in expression and their diagnostic value.
Random Forest (RF) and Support Vector Machine (SVM) Algorithm
The Random Forest structure is utilized to determine the mean decrease impurity, commonly referred to as the Gini significance. Decision Trees are part of Random Forest. Every decision tree has internal nodes and leaves. The internal node decides how to divide the data set into two subgroups with related responses using the selected characteristic. For classification or information gain tasks, gini impurity or information gain criteria may be utilized, while variance reduction may be used for regression activities. Each feature’s impact on the impurity of the split can be measured (the feature with highest decrease is selected for internal node). We can monitor the average impurity decrease for each feature. The relative importance of a feature is determined by taking the average of all the trees in the forest. The Scikit-Learn Random Forest implementation provides this method (for both classifier and regressor). SVM was employed to evaluate the diagnostic value of specific genes taken together.
Detection of Immune Infiltrating Cells in EC and EU Endometriosis
The relative proportions of 22 immunological components were calculated using the CIBERSORT algorithm. A gene expression matrix and particular gene sets from 22 different types of immune cells were used in the simulation calculation, which was carried out 1000 times. These immune cells’ respective composition ratios in each tissue were calculated. Using the ESTIMATE algorithm, immune and stromal scores for each tissue were calculated.
Human Tissue Collection
The tissue samples in this study were obtained from 8 patients with EC and EU tissues paraffin block from post-operation patients with EMT in Anhui Maternal and Child Health Hospital from January 2020 to October 2022.
Immunohistochemical Examination (IHC)
EC and EU endometrium were collected, and paraffin sections were made by dewaxing, antigen repair and sealing, and then incubated overnight at 4°C with HRP-labeled polyclonal antibodies against CXCL12, PECAM1, NGF, CTGF and WNT5A. Goat anti-rabbit secondary antibody was incubated at room temperature for 50 min, and freshly prepared DAB color development solution was added dropwise, and the color development time was controlled under the microscope. Then, the results of CXCL12, PECAM1, NGF, CTGF and WNT5A staining in each tissue sample were quantified using Image Pro-Plus 6.0 software, and the average optical density (AOD) values were calculated.
Statistical Analysis
Cytoscape (version 3.9.1) and R (version 4.2.1) were utilized for the statistical analysis in this work. The bilateral test also had a P<0.05 statistical significance level. For determining the differences in variables among groups, the Student’s t-test, Kruskal–Wallis, Wilcoxon, or one-way ANOVA test was used. The Area Under Curve (AUC) values were used to determine each remarkable gene’s predictive potential. The correlation test Spearman and Pearson were applied to determine the associations between variables.
Results
Identify Differential Genes (DEGs)
We used high-throughput sequencing technology to perform whole-transcriptome sequencing on EC and EU samples from GSE141549 in order to investigate the pathophysiology of EMT. Firstly, principal component analysis (Figure 1A) was utilized to examine whether there was a difference between the EU and EC using “factoextra” tools based on sequencing data. Using “limma” packages, we also found 926 DEGs in EC group compared to EU group. 375 genes were down-regulated, and 551 up-regulated (Figure 1B).
WGCNA Analysis the Most Relevant to Endometriosis Module
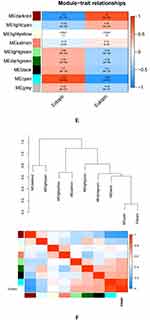
To further explore genes related to EMT, we utilized the GSE141549 dataset and performed WGCNA. Genes with the top 10,000 median absolute values were filtered to construct the WGCNA network. Cluster analysis excludes samples with obvious outliers and erroneous clustering (Figure 2A). The remaining 218 samples were analyzed for the next step (Figure 2B). Choose 11 as the optimal soft threshold, R2 = 0.9 (Figure 2C). In accordance with the dynamic tree species cutting method, the genes were separated into several modules based on the appropriate soft threshold. After merging, a total of nine modules (Figure 2D), including the dark-red module (3157 genes), the light-cyan module (215 genes), the light-yellow module (493 genes), the salmon module (1580 genes), the light-green module (143 genes), the dark-green module (497 genes), the black module (423 genes), the cyan module (1721 genes), and the grey module (1771 genes) were produced. And then, we calculated the module’s association with the clinical features. As shown in Figure 2E and F, the largest positive link with the EC was found in the cyan module.(cor = 0.86, P = 3 e–66). To determine the relationship between the color module and gene significance, a thorough calculation was done. The cyan module was closely related to EC (cor=0.9, P<1e-200; Figure 2G). Next, we will analyze the cyan module’s subsequent step.
Figure 2 Continued. Figure 2 Continued. Figure 2 Continued.

Screening for AID-DEGs Associated with Endometriosis
Through the screening and intersection of genes in the cyan module (n=1721), gene sets downloaded from the GAAD (n=4186) and DEGs (n=926) according to gene names, a total of 114 genes related to EMT associated with Autoimmune diseases (AID-DEGs) were obtained. The results shown in the Venn diagram above (Figure 3).
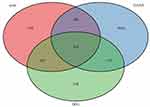 |
Figure 3 Screening of autoimmune gene-related genes for key module genes involved in the development of endometriosis. |
Function and Pathway Analysis of AID-DEGs Associated with Endometriosis
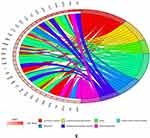
Positive regulation of “response to external stimulus”, “leukocyte migration”, “regulation of actin cytoskeleton organization”, “regulation of leukocyte chemotaxis”, “transmembrane receptor protein serine/threonine kinase signaling pathway”, “positive regulation of leukocyte migration”, “cellular extravasation”, and “regulation of angiogenesis” were all enriched in the 114 AID-DEGs. GO biological processes (Figure 4B). In molecular function (MF), “receptor ligand activity” and “signaling receptor activator activity” were the most significantly enriched pathways. “Collagen containing extracellular matrix” pathways were prominent in the cellular component (CC) pathways (Figure 4A). The AID-DEGs were shown to participate in the pathways “Cell adhesion molecules”, “Cytokinecytokine receptor interaction”, and “Leukocyte transendothelial migration”, according to KEGG pathway analysis (Figure 4D–E). Genes assigned to the top 6 biological process GO terms are displayed in (Figure 4C).
Figure 4 Continued. Figure 4 Continued.
PPI Network and Identifying Hub Genes
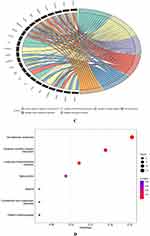
The STRING online database (version 11.5) was used to import 114 AID-DEGs in total to create the PPI network. Cytoscape’s (version 3.9.1) processing of the protein network is displayed in (Figure 5A). The Cytohubba (version 0.1) plug-ins of Cytoscape were used to assess the 114 genes with interactions using five different types of algorithms (MCC, MNC, Degree, EPC, and Betweenness) (Table 1). A Venn diagram displaying the top 10 genes identified by each algorithm is depicted in Figure 5B. Five hub genes were identified from the overlapped region are Connective Tissue Growth Factor (CTGF), C-X-C Motif Chemokine Ligand 12 (CXCL12), Nerve Growth Factor (NGF), Platelet And Endothelial Cell Adhesion Molecule 1(PECAM1), Wnt family member 5A (WNT5A).
 |
Table 1 Top 10 Essential Genes for 5 Algorithms (MCC, MNC, Degree, EPC, and Betweenness) |
 |
Figure 5 Hub genes of AID-DEGs were selected. (A) Protein interaction analysis of 114 genes using the STRING online website. (B) Venn diagram intersection of the top 10 genes. |
Analysis and Exploration Differences in the Expression and the Diagnostic Value of Hub Genes
The differences between the expression of hub genes in the EU and EC were examined using the GSE141549 dataset. As illustrated in Figure 6A, the expression levels of CTGF (P<2.2e-16), CXCL12 (P<2.2e-16), NGF (P<2.2e-16) and PECAM1 (P<2.2e-16) were higher in EC than in EU. We analysed the expression of hub genes in EC and EU tissues in different types of endometriosis and found that the expression of hub genes in EC tissues in different types of endometriosis was consistent relative to the endometrium, as shown in Supplementary Figure 1. In particular, EU exhibits higher levels of WNT5A (P<1.9e-14) expression than EC. As shown in Figure 6D, the expression of hub genes in GSE7305 was the same as GSE141549. Finally, using the “pROC” R package, this work evaluated the diagnostic value of the hub genes in EMT with GSE141549, with the following results (Figure 6B): WNT5A:AUC=0.8215, CTGF:AUC=0.9048, NGF:AUC=0.9318. PECAM1:AUC=0.9295, CXCL12:AUC=0.9327. By displaying ROC curves, the diagnostic value of five hub genes was confirmed (Figure 6C), the result as follows: PECAM1: AUC=0.97, CXCL12:AUC=0.99, WNT5A:AUC=0.82, CTGF:AUC=0.79, NGF:AUC=1. The validation results were consistent with the analysis results, and the hub genes was expressed differently in EMT and had diagnostic value.
Figure 6 Continued.
Results of Diagnostic Value Testing of Hub Genes via RF and SVM Algorithms
The gini importance of the random forest was used to compare the importance of CXCL12, PECAM1, NGF, CTGF, and WNT5A for the diagnosis of the EMT, as shown in the figure (Figure 6E), CXCL12: MeanDecreaseGini=26.127; PECAM1: MeanDecreaseGini=20.291; NGF: MeanDecreaseGini=24.110;CTGF:MeanDecreaseGini=11.792; WNT5A:MeanDecreaseGini = 7.766. Using the SVM algorithm, the 236 samples were divided into the training set and the test set according to the ratio of the train: test 7:3, and the joint diagnosis model of hub genes for EMT was constructed, and the ROC curve was drawn (Figure 6F) AUC=0.945.
Relationship Between AID-DEGs Hub Genes and Immune Infiltrating Cells in EC and EU
Immune traits were evaluated in relation to immune-checkpoint, HLA, receptor, chemokine, and abundance levels of immune cells. When compared to the EU group, the levels of the majority of immunological checkpoints, HLAs, receptors, and chemokines are higher in the EC group (Figure 7A). We determined the proportions of 22 immunological components between EC and EU endometrial tissues using the CIBERSORT method (Figure 7B). Figure 7C showed the intricate interactions between these immune compositions, with resting macrophages and mast cells particularly associated with the majority of immune compositions. Additionally, compared to EU, most immune cells were more abundant in EC endometrium tissues. The five hub genes CXCL12, PECAM1, NGF, and CTGF were positively correlated with monocytes, macrophages, B cell memory, mast cells, and negatively correlated with regulatory T cells and NK cells, according to further research. WNT5A was substantially connected with regulatory T cells and NK cells, but was not adversely correlated with monocytes, macrophages, or mast cells (Figure 7D).
Figure 7 Continued.
Hub Genes Expression in EC and EU Tissues
The results of immunohistochemical staining revealed that CXCL12 and CTGF were mainly expressed in the cytoplasm and mainly localized in endometrial glands and mesenchymal cells; WNT5A was mainly located in the cytoplasm of glands; PECAM1 was mainly located in the cytoplasm of vascular endothelial cells and mesenchymal cells; NGF was mainly located in the cytoplasm of endometrial glands (Figure 8A). The staining result of hub genes in EC and EU endometrium was analyzed by image pro plus software, and the AOD values were calculated. CXCL12, NGF, CTGF, and PECAM1 showed higher AOD in EC endometrium than in orthotopic endometrium (p<0.05) (Figure 8B), while WNT5A showed low expression in EC endometrium.
Figure 8 Continued. Figure 8 Continued.

Hub Genes Expression in Menstrual Cycle
We analyzed the relationship between the screened hub genes and menstrual cycle and found no significant difference in the expression of CXCL12, CTGF, NGF and WNT5A during the menstrual, proliferative and secretory phases, indicating that the menstrual cycle had no effect on the expression of CXCL12, CTGF, NGF and WNT5A; while the expression of PECAM1 was higher in the menstrual phase than in the secretory and proliferative phases, as shown in the Supplementary Figure 2.
Discussion
EMT is a non-malignant gynecological illness with an uncertain mechanism. Laparoscopy is still the gold standard for diagnosis EMT, and no particular treatment targets or biomarkers have been found.29,30 Short menstrual cycles, low body mass index, low waist-to-hip ratio, and low gestational age are risk factors for endometriosis, but studies have also found an increased risk of melanoma, non-Hodgkin’s lymphoma, thyroid cancer, and endometrial cancer in endometriosis.31 The etiology of EMT is not known, but theories about it include the “implantation theory” and “existing endometrial determinism”. The pathophysiology of EMT is not known, but it is widely believed to be related to immunity, genetics, retrograde menstruation, and lymphatic blood. A meta-analysis showed that women with endometriosis are at increased risk of several autoimmune diseases and that EMT appears to be associated with autoimmune diseases of the body.32
Autoimmune diseases33 refers to the body to antigen immune response to their organs or tissue damage caused by a kind of disease, can be divided into systemic autoimmune diseases (such as rheumatoid arthritis, systemic lupus erythematosus) and organ-specific autoimmune diseases (such as chronic lymphocytic thyroiditis, hyperthyroidism, ulcerative colitis, mainly according to the typical clinical manifestations and related examination diagnosis, and the high incidence of women, the course of delay. These are primarily determined by the typical clinical manifestations and associated examination diagnosis It has been demonstrated in animal models that immunosuppressants such as thalidomide and triptolide, as well as biological agents such as etanercept and infliximab, are beneficial in reducing the size of ectopic lesions and EMT in the treatment of autoimmune disorders.34,35
Through this study, we screened out the AIDGs hub genes related to EMT, CXCL12, PECAM1, NGF, CTGF, and WNT5A, and analyzed the diagnostic value of hub genes and the correlation with immune microenvironment. First, by analyzing the immune microenvironment of the EU and EC endometrium, it was found that macrophages, mast cells and T cells CD4 memory quiescent in the EC endometrium had a high percentage, while NK cells and plasma cells accounted for a relatively small percentage. By investigating the relationship between the hub genes and the immune environment, we found that CXCL12, PECAM1 and NGF were all strongly associated with mast cells, macrophages and activated NK cells, respectively. CXCL12, PECAM1 and NGF were highly expressed in EC endometrium. Therefore, we speculate that CXCL12, PECAM1 and NGF may promote EMT by activating macrophages, mast cells and inhibiting NK cell activation. In contrast, WNT5A was lowly expressed in EC endometrium and was positively correlated with NK cells according to immune infiltration analysis. It is hypothesized that NK cells are absent in EC endometrium due to the low expression of WNT5A.
CXCL12, an essential homeostatic chemokine, is also referred as stromal cell-derived factor-1 (SDF-1).36 In physiological and pathological processes such as embryogenesis, hemocyogenesis, angiogenesis, and inflammation, CXCL12 plays a critical role.37 Hematopoietic progenitor cells, stem cells, endothelial cells, and the majority of leukocytes can all be activated and induced to migrate.38 The CXCL12/CXCR 4 axis is extensively expressed in EMT, and activation of the AKT pathway through the CXCL12/CXCR 4 axis promotes the invasiveness of uterine EC endometrial cells and the development of ectopic focal blood vessels.39 PECAM-1 (CD31)40 can be identified on a range of cells in the vascular compartment.41 Due to its involvement in numerous mechanisms, including leukocyte movement through intracellular junctions, cell death, angiogenesis, etc, PECAM-1 plays a variety of diverse and broad roles in inflammation.42 Inhibiting PECAM1 in animal models of arthritis can stop the advancement of inflammation. Additionally, it has been discovered that through stimulating the NF-κB signaling pathway, PECAM1 and CXCR 4 can interact to increase the inflammatory response in pulpitis samples.43 A literature review found that NGF is associated with severe pain in EMT.44 Due to the recruitment and polarization of macrophages in EMT tissue. Macrophages express different nerve growth factors (NGF) and NGF causes neurogenic imbalance in an estrogen-dependent manner.45 Patients with EMT have neuropathic pain, which is a result of estrogen-controlled neuroimmune interactions, which sensitize peripheral neurons. CTGF is a diagnostic and therapeutic target for rheumatoid arthritis (RA) due to its role in cell adhesion, fibrosis, and angiogenesis in the early stages of the disease.46,47 Accordingly, it is presumed that CTGF may influence EMT to transform from a stromal fibroblast phenotype to a myofibroblast-like phenotype, which is responsible for the characteristics of EMT.48 WNT5A is an example of a ligand that can control cell motility and polarity by signal transduction and can also function in macrophages as a pro-inflammatory agent to cause inflammation.49 It was discovered to be expressed in a number of autoimmune disorders and was linked to disease development, such as systemic lupus erythematosus studies that linked the expression of WNT5A protein to disease progression.50 It had discovered that WNT5A can transmit RYK / RhoA / ROCK signaling and WNT / Ca 2 + expression through inflammatory mediators, which is favorable to RA migration and invasion.51 Recent research demonstrates that WNT5A reduces the development of monocytes into inflammatory M1 macrophages and generates an immunosuppressive macrophage phenotype. This is consistent with what we discovered.52
Admittedly, there are some limitations in this study. First, we did not validate the immune microenvironment components of EC and EU endometrium in vivo. Second, the correlation between hub genes and immune microenvironment was also not validated in vivo. Therefore, in the follow-up study, we will continue to explore the association of hub genes with endometriosis severity and pharmacological treatment, which will also allow us to explore new ideas about the pathogenesis and immunotherapy of EMT.
Conclusion
In conclusion, in our study, bioinformatics and experimental method were used to identify hub genes (CXCL12, PECAM1, NGF, CTGF, and WNT5A) for autoimmune diseases associated with the immune microenvironment of EMT. Data analysis and in vitro experiments confirmed that hub genes are differentially expressed in EU and EC. Furthermore, hub genes are important for the diagnosis of EMT and the immune microenvironment, although our study also has some limitations. However, we must pay more attention to hub genes and further investigate the molecular mechanisms underlying the role of hub genes in EMT and autoimmune diseases, as well as their immunotherapeutic effects on EMT, to facilitate further development of immunotherapy in EMT.
Abbreviations
EMT, endometriosis; AID, autoimmune diseases; AIDGs, autoimmune disease related genes; MAF, mutant allele frequencies; KRAS, Kirsten rats arcomaviral oncogene homolog; TPOAb, thyroid peroxidase antibodies; WGCNA, Weighted gene co-expression network analysis; GEO, Gene Expression Omnibus; TOM, topological overlap matrix; DEGs, differentially expressed genes; AID-DEGs, autoimmune disease gene-related differential genes; EC, ectopic endometrium; EU, eutopic endometrium; BP, biological process; MF, molecular function; CC, cellular component; RF, Random forest; SVM, Support Vector Machine; ROC, Receiver operating characteristic curve; AUC, The area under the Curve; GO, Gene Ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes; PPI, Protein- Protein Interaction network; IHC, Immunohistochemical examination; CTGF, Connective Tissue Growth Factor; CXCL12, C-X-C Motif Chemokine Ligand 12; NGF, Nerve Growth Factor; PECAM1, Platelet And Endothelial Cell Adhesion Molecule 1; WNT5A, Wnt family member 5A.
Data Sharing Statement
We conducted this study using public databases, and all of the data from the above studies are available in the database:GEO (http://www.ncbi.nlm.nih.gov/geo), GAAD database (http://gaad.medgenius.info/genes/).
Ethics Approval
The study complies with the Declaration of Helsinki. The Anhui Province Maternity and Child Healthcare Hospital Ethics Committee approved the study protocol (Ethical number:YYLL2019-2019xjk173-01-01). All participants were informed of the study procedures and objectives, and this study was conducted after participants signed an informed consent form.
Informed Consent
Patients and their family members could actively engage in the trial. On the basis that they have a complete comprehension of the therapeutic approach, all patients will sign the “informed consent form” and offer informed consent to the trial process. Maximizing the protection of the participants’ rights, interests, safety, and health is the basis of the clinical trial protocol.
Acknowledgments
We appreciate the authors who uploaded their valuable datasets to the GEO and GAAD databases for donating their platforms. Many thanks also to the patients and families who were included in the study. Yin-Ting Yang, Xi-Ya Jiang, Hong-Liang Xu, and Guo Chen are co-first authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The Natural Science Foundation of Higher Education Institutions of Anhui Province (Grant No. KJ2021A0352, Grant No. YJS20230095), the Research Fund Project of Anhui Medical University (Grant No. 2020xkj236), and the Applied Medicine Research Project of Hefei Health Commission all provided funding in support of this work.
Disclosure
The authors affirm that there were no financial or commercial ties that might be considered to be a potential conflict of interest for this work.
References
1. Hung SW, Zhang R, Tan Z, Chung J, Zhang T, Wang CC. Pharmaceuticals targeting signaling pathways of endometriosis as potential new medical treatment: a review. Med Res Rev. 2021;41:2489–2564. doi:10.1002/med.21802
2. Parazzini F, Esposito G, Tozzi L, Noli S, Bianchi S. Epidemiology of endometriosis and its comorbidities. Eur J Obstet Gynecol Reprod Biol. 2017;209:3–07. doi:10.1016/j.ejogrb.2016.04.021
3. As-Sanie S, Black R, Giudice LC, et al. Assessing research gaps and unmet needs in endometriosis. Am J Obstet Gynecol. 2019;221(2):86–94. doi:10.1016/j.ajog.2019.02.033
4. Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med. 2020;382:1244–1256. doi:10.1056/NEJMra1810764
5. Sampson JA. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am J Pathol. 1927;3:93–110.
6. Lang J. Cornerstone of study on endometriosis (in Chinese). Chin J Obstet Gynecol. 2005;40(1):3–4.
7. Zhang T, De Carolis C, Man G, Wang CC. The link between immunity, autoimmunity and endometriosis: a literature update. Autoimmun Rev. 2018;17:945–955. doi:10.1016/j.autrev.2018.03.017
8. Ahn SH, Monsanto SP, Miller C, Singh SS, Thomas R, Tayade C. Pathophysiology and immune dysfunction in endometriosis. Biomed Res Int. 2015;2015:795976. doi:10.1155/2015/795976
9. Suda K, Nakaoka H, Yoshihara K, et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24(7):1777–1789. doi:10.1016/j.celrep.2018.07.037
10. Jin Z, Zhang Y, Li J, Lv S, Zhang L, Feng Y. Endometriosis stem cell sources and potential therapeutic targets: literature review and bioinformatics analysis. Regen Med. 2021;16(10):949–962. doi:10.2217/rme-2021-0039
11. Lac V, Nazeran TM, Tessier-Cloutier B, et al. Oncogenic mutations in histologically normal endometrium: the new normal? J Pathol. 2019;249:173–181. doi:10.1002/path.5314
12. Yin Z, Low HY, Chen BS, et al. Risk of ankylosing spondylitis in patients with endometriosis: a population-based retrospective cohort study. Front Immunol. 2022;13:877942. doi:10.3389/fimmu.2022.877942
13. Porpora MG, Scaramuzzino S, Sangiuliano C, et al. High prevalence of autoimmune diseases in women with endometriosis: a case-control study. Gynecol Endocrinol. 2020;36(4):356–359. doi:10.1080/09513590.2019.1655727
14. Abramiuk M, Grywalska E, Malkowska P, Sierawska O, Hrynkiewicz R, Niedzwiedzka-Rystwej P. The role of the immune system in the development of endometriosis. Cells. 2022;11. doi:10.3390/cells11132028
15. Yuk JS, Park EJ, Seo YS, Kim HJ, Kwon SY, Park WI. Graves disease is associated with endometriosis: a 3-year population-based cross-sectional study. Medicine. 2016;95(10):e2975. doi:10.1097/MD.0000000000002975
16. Sun YH, Leong PY, Huang JY, Wei JC. Increased risk of being diagnosed with endometriosis in patients with systemic lupus erythematosus: a population-based cohort study in Taiwan. Sci Rep. 2022;12:13336. doi:10.1038/s41598-022-17343-4
17. Kvaskoff M, Mu F, Terry KL, et al. Endometriosis: a high risk population for major chronic diseases? Hum Reprod Update. 2015;21:500–516. doi:10.1093/humupd/dmv013
18. Chao YH, Liu CH, Pan YA, Yen FS, Chiou JY, Wei JC. Association between endometriosis and subsequent risk of Sjogren’s syndrome: a nationwide population-based cohort study. Front Immunol. 2022;13:845944. doi:10.3389/fimmu.2022.845944
19. Gabriel M, Fey V, Heinosalo T, et al. A relational database to identify differentially expressed genes in the endometrium and endometriosis lesions. Sci Data. 2020;7:284. doi:10.1038/s41597-020-00623-x
20. Hever A, Roth RB, Hevezi P, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc Natl Acad Sci USa. 2007;104:12451–12456. doi:10.1073/pnas.0703451104
21. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi:10.1093/nar/gkv007
22. Maag J. Gganatogram: an R package for modular visualisation of anatograms and tissues based on ggplot2. F1000Res. 2018;7:1576. doi:10.12688/f1000research.16409.2
23. Wu T, Hu E, Xu S, et al. ClusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi:10.1016/j.xinn.2021.100141
24. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi:10.1093/nar/gkaa1074
25. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
26. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4(1):2. doi:10.1186/1471-2105-4-2
27. Chin CH, Chen SH, Wu HH, et al. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
28. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi:10.1186/1471-2105-12-77
29. Matalliotakis IM, Cakmak H, Fragouli YG, Goumenou AG, Mahutte NG, Arici A. Epidemiological characteristics in women with and without endometriosis in the Yale series. Arch Gynecol Obstet. 2008;277(5):389–393. doi:10.1007/s00404-007-0479-1
30. Farland LV, Eliassen AH, Tamimi RM, Spiegelman D, Michels KB, Missmer SA. History of breast feeding and risk of incident endometriosis: prospective cohort study. BMJ. 2017;358:j3778. doi:10.1136/bmj.j3778
31. Schernhammer ES, Vitonis AF, Rich-Edwards J, Missmer SA. Rotating nightshift work and the risk of endometriosis in premenopausal women. Am J Obstet Gynecol. 2011;205:471–476. doi:10.1016/j.ajog.2011.06.002
32. Shigesi N, Kvaskoff M, Kirtley S, et al. The association between endometriosis and autoimmune diseases: a systematic review and meta-analysis. Hum Reprod Update. 2019;25(4):486–503. doi:10.1093/humupd/dmz014
33. Xiao ZX, Miller JS, Zheng SG. An updated advance of autoantibodies in autoimmune diseases. Autoimmun Rev. 2021;20(2):102743. doi:10.1016/j.autrev.2020.102743
34. Kotlyar A, Taylor HS, D’Hooghe TM. Use of immunomodulators to treat endometriosis. Best Pract Res Clin Obstet Gynaecol. 2019;60:56–65. doi:10.1016/j.bpobgyn.2019.06.006
35. Antonio L, Rosa-E-Silva JC, Machado DJ, et al. Thalidomide reduces cell proliferation in endometriosis experimentally induced in rats. Rev Bras Ginecol Obstet. 2019;41:668–672. doi:10.1055/s-0039-3399551
36. Liepelt A, Tacke F. Stromal cell-derived factor-1 (SDF-1) as a target in liver diseases. Am J Physiol Gastrointest Liver Physiol. 2016;311(2):G203–G209. doi:10.1152/ajpgi.00193.2016
37. Moridi I, Mamillapalli R, Cosar E, Ersoy GS, Taylor HS. Bone marrow stem cell chemotactic activity is induced by elevated CXCl12 in endometriosis. Reprod Sci. 2017;24(4):526–533. doi:10.1177/1933719116672587
38. Barbieri F, Bajetto A, Thellung S, Wurth R, Florio T. Drug design strategies focusing on the CXCR4/CXCR7/CXCL12 pathway in leukemia and lymphoma. Expert Opin Drug Discov. 2016;11(11):1093–1109. doi:10.1080/17460441.2016.1233176
39. Mei J, Li MQ, Li DJ, Sun HX. MicroRNA expression profiles and networks in CXCL12‑stimulated human endometrial stromal cells. Mol Med Rep. 2017;15:249–255. doi:10.3892/mmr.2016.5997
40. Simmons DL, Walker C, Power C, Pigott R. Molecular cloning of CD31, a putative intercellular adhesion molecule closely related to carcinoembryonic antigen. J Exp Med. 1990;171:2147–2152. doi:10.1084/jem.171.6.2147
41. Newman PJ, Newman DK. Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler Thromb Vasc Biol. 2003;23:953–964. doi:10.1161/01.ATV.0000071347.69358.D9
42. Stockinger H, Gadd SJ, Eher R, et al. Molecular characterization and functional analysis of the leukocyte surface protein CD31. J Immunol. 1990;145(0022–1767):3889–3897.
43. Liu Y, Zhang Z, Li W, Tian S. PECAM1 combines with CXCR4 to trigger inflammatory cell infiltration and pulpitis progression through activating the NF-kappaB signaling pathway. Front Cell Dev Biol. 2020;8:593653. doi:10.3389/fcell.2020.593653
44. Peng B, Zhan H, Alotaibi F, Alkusayer GM, Bedaiwy MA, Yong PJ. Nerve growth factor is associated with sexual pain in women with endometriosis. Reprod Sci. 2018;25(4):540–549. doi:10.1177/1933719117716778
45. Liang Y, Xie H, Wu J, Liu D, Yao S. Villainous role of estrogen in macrophage-nerve interaction in endometriosis. Reprod Biol Endocrinol. 2018;16(1):122. doi:10.1186/s12958-018-0441-z
46. Wu G, Liu C, Cao B, et al. Connective tissue growth factor-targeting DNA aptamer suppresses pannus formation as diagnostics and therapeutics for rheumatoid arthritis. Front Immunol. 2022;13:934061. doi:10.3389/fimmu.2022.934061
47. Li D, Finley SD. Exploring the extracellular regulation of the tumor angiogenic interaction network using a systems biology model. Front Physiol. 2019;10:823. doi:10.3389/fphys.2019.00823
48. Kusama K, Fukushima Y, Yoshida K, et al. PGE2 and thrombin induce myofibroblast transdifferentiation via activin A and CTGF in endometrial stromal cells. Endocrinology. 2021:162. doi:10.1210/endocr/bqab207
49. Awan S, Lambert M, Imtiaz A, et al. Wnt5a promotes lysosomal cholesterol egress and protects against atherosclerosis. Circ Res. 2022;130(2):184–199. doi:10.1161/CIRCRESAHA.121.318881
50. Chi S, Xue J, Chen X, Liu X, Ji Y. Correlation of plasma and urine Wnt5A with the disease activity and cutaneous lesion severity in patients with systemic lupus erythematosus. Immunol Res. 2022;70:174–184. doi:10.1007/s12026-021-09253-w
51. Rodriguez-Trillo A, Pena C, Garcia S, et al. ROCK inhibition with Y-27632 reduces joint inflammation and damage in serum-induced arthritis model and decreases in vitro osteoclastogenesis in patients with early arthritis. Front Immunol. 2022;13:858069. doi:10.3389/fimmu.2022.858069
52. Sumitomo R, Huang CL, Ando H, Ishida T, Cho H, Date H. Wnt2b and Wnt5a expression is highly associated with M2 TAMs in non‑small cell lung cancer. Oncol Rep. 2022;48. doi:10.3892/or.2022.8404
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.