Back to Journals » Cancer Management and Research » Volume 12
ATP6AP2 is Overexpressed in Breast Cancer and Promotes Breast Cancer Progression
Authors Zhao K , Wang M , Wu A
Received 9 July 2020
Accepted for publication 14 September 2020
Published 22 October 2020 Volume 2020:12 Pages 10449—10459
DOI https://doi.org/10.2147/CMAR.S270024
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Kankan Zhao,* Mengchuan Wang,* Aiguo Wu
Department of General Surgery, Zhujiang Hospital, The Second School of Clinical Medicine, Southern Medical University, Guangzhou 510282, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Aiguo Wu Email [email protected]
Background: Adenosine triphosphatase H+ transporting accessory protein 2 (ATP6AP2), also known as (pro)renin receptor, is implicated in tumorigenesis and the progression of several types of cancer. This study investigated the role of ATP6AP2 in breast cancer.
Methods: UALCAN and ONCOMINE datasets were utilized to compare transcript levels of ATP6AP2 in breast cancer and normal tissues. GOBO datasets were applied to examine ATP6AP2 expression in different breast cancer cell lines. We used the cBioPortal website to explore the gene alterations and copy number alterations of ATP6AP2 in breast cancer. Cell Counting Kit-8 and transwell assays were conducted to evaluate ATP6AP2 function in MCF-7 breast cancer cells. Finally, we used the cBioPortal website to establish the interaction network of ATP6AP2 in breast cancer and performed functional enrichment analysis based on Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways.
Results: ATP6AP2 was overexpressed in breast cancer tissues and breast cancer cell lines in the UALCAN, ONCOMINE, and GOBO datasets. The major type of ATP6AP2 alteration was mRNA upregulation. Moreover, ATP6AP2 was most highly expressed in luminal type breast cancer. Finally, ATP6AP2 knockdown reduced MCF-7 cell proliferation, invasion and migration. Functional enrichment analysis suggested that ATP6AP2 regulates several cancer-related pathways, especially the Wnt/β-catenin signaling pathway.
Conclusion: Applying multi-dimensional analytical methods, we demonstrate that ATP6AP2 is upregulated in breast cancer and may promote its development and progression.
Keywords: breast cancer, ATP6AP2, PRR, bioinformatics analysis
Introduction
Breast cancer is common all over the world and the incidence is continuously increasing. According to the statistics of International Agency for Research on Cancer (IARC), 567,233 new cases of breast cancer occurred worldwide in 2018, accounting for 5.1% of all cancer types in women.1 Although the molecular mechanisms underlying breast tumorigenesis have been well studied in recent years, the prognosis for different breast cancer types is still unsatisfactory. Currently, cancer-targeting therapies are promising treatments for cancer. These therapies target at blocking the growth and spread of cancer by interfering with specific molecules. However, these molecules which are involved in the growth, progression and metastasis of breast cancer are limited. Thus, it is of great importance to find novel biomarkers contribute to clinical decision-making.
Adenosine triphosphatase H+ transporting accessory protein 2 (ATP6AP2), also known as (pro)renin receptor (PRR), encodes a protein associated with adenosine triphosphatases (ATPases),2 which play important roles in energy conservation, secondary active transport, acidification of intracellular compartments, and cellular homeostasis. ATP6AP2 is able to interact with several signaling pathways and molecules which include mitogen-activated protein kinase pathway, extracellular signal-regulated kinase 1 and 2, phosphoinositide 3 kinase, transforming growth factor β1, and Wnt/β-catenin signaling.3 Previous studies have shown that ATP6AP2 is implicated in tumorigenesis and the progression of several types of cancer.4–7 One study with 69 sample sizes found that ATP6AP2 was upregulated in breast cancer tissues and could trigger proliferation of breast cancer cells.8 However, other functions of ATP6AP2 were not explored and the mechanism is unclear. As ATP6AP2 is associated with several important pathways related to cancer, the role of ATP6AP2 in breast cancer deserves further research.
With the development of microarray and deep sequencing technology, biological and biomedical studies have been revolutionized.9 There are thousands of gene expression data online, which provide an easy access to retrieving and exploiting these data. Therefore, in our study, we combined bioinformatics analysis and experimental approaches to explore the role of ATP6AP2 in breast cancer.
Materials and Methods
UALCAN Analysis
The UALCAN website (http://ualcan.path.uab.edu) is a comprehensive web resource for analyzing cancer omics data.10 We used the website to examine ATP6AP2 mRNA expression in breast cancer. We also evaluated the associations of ATP6AP2 expression with clinicopathologic characteristics of breast cancer patients.
ONCOMINE Analysis
ONCOMINE is an online cancer microarray database (www.oncomine.org), allowing us to explore the expression difference of genes between cancer samples and normal samples.11 To validate the results obtained from the ULCAN dataset, we further explored the expression of ATP6AP2 in breast cancer using this database.
Gene Expression-Based Outcome for Breast Cancer Online (GOBO) Analysis
GOBO (http://co.bmc.lu.se/gobo/) is an online tool that allows rapid assessment of gene expression levels in an 1881-sample breast cancer data set.12 In addition, GOBO allows us to investigate gene expression levels in breast cancer subtypes and breast cancer cell lines. We analyzed ATP6AP2 transcript levels in breast cancer cell lines by using GOBO database.
Cell Culture and Si-RNA Transfection
MCF-7 breast cancer cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in Roswell Park Memorial Institute 1640 medium (Gibco, USA), containing 10% heat-inactivated fetal bovine serum at 37°C in a humidified 5% CO2 atmosphere. Human PRR small interfering (si)RNA (target sequences: GGATCATTCTCCTGATTTA) and non-targeting control siRNA (NC-siRNA) were designed and synthetized by Ribobio company (Guangzhou, China). MCF-7 cell lines were transfected with either human PRR siRNA (20 μmol) or NC-siRNA (20 μmol) as controls for 48h by using the LipofectamineTM 3000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Then, the effect of the siRNA transfection was verified by reverse transcription PCR, and cells were collected for further analysis.
RNA Extraction and Quantitative Real-Time (qRT)-PCR
Total RNA was extracted using TRIzol reagent (Sigma, St. Louis, MO, USA), and reverse transcription reactions were performed using an All-in-One synthesis kit (GeneCopoeia, USA) according to manufacturers’ recommendations. qRT-PCR was performed on a CFX PCR detection system (Bio-Rad, Hercules, USA). Relative gene expression levels were calculated using the ΔΔCt method and normalized to that of the GAPDH. Primers used for qRT-PCR are listed in Supplemental Materials.
Western Blotting
MCF-7 breast cancer cells were homogenized and lysed in RIPA buffer containing protease and phosphatase inhibitors (cOmplete™ Protease Inhibitor Cocktail, Roche, Shanghai, China). We measured the protein concentration of the homogenized sample using a bicinchoninic acid assay kit (Invitrogen), according to the manufacturer’s recommendations. The samples were boiled for 10min in a water bath. We separated the protein in the extracts using 10%SDS-PAGE and transferred them to nitrocellulose membranes. Subsequently, membranes were blocked with 3% BSA, washed in 1×TBST and incubated overnight with primary antibodies. The primary antibodies are listed in Supplemental materials. The blots were scanned using an Infrared Imaging System (Odyssey, LI-COR) and quantitatively analyzed using Image J software.
Cell Growth Assay
Breast cancer cell growth ability was assessed by Cell Counting Kit-8 assay (CCK-8, Dojindo, Japan). The cells were suspended in complete medium 24, 48, or 72 h after transfection and seeded into a 96-well plate at a concentration of 1× 104 cells (100 μL) per well. Absorption (optical density [OD] value) was measured at 450 nm using a microplate spectrophotometer (Molecular Devices, USA).
Cell Invasion and Migration Assay
For migration and invasion assays, 8 µm pore size transwell inserts (BD Biosciences, USA) were used. Uncoated and Matrigel-coated transwell inserts were used to examine MCF-7 cell migration and invasion, respectively. We added 1×105 cells in 100 µL to the upper chamber and 500 µL complete medium to the bottom chamber. Migrating or invading cells were washed and fixed in 4% paraformaldehyde for 10 min, followed by crystal violet indicator (Sigma, USA) staining. Five visual fields were selected randomly using an IX53 inverted microscope (Olympus, Japan) at 200x magnification.
Functional Enrichment Analysis
The cBio Cancer Genomics Portal (cBioportal) (http://www.cbioportal.org) is an online platform for exploring cancer genomics data.13 The cBioPortal breast invasive carcinoma datasets (TCGA, PanCancer Atlas) containing 1084 samples were selected to explore the gene alterations and functions of ATP6AP2 in breast cancer. Using cBioportal, we constructed a network to show ATP6AP2 in the context of biological interactions derived from public pathway databases. Next, we applied clusterProfiler package14 in R language to perform Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis on the genes in the network. GO terms of biological process (BP), cellular components (CC) and molecular functions (MF), and KEGG pathways with a false discovery rate <0.05 were considered significant and visualized as bubble diagrams.
Statistical Analysis
All experiments were repeated three times. Statistical analysis was performed using GraphPad Prism (GraphPad Software Inc, San Diego, CA). Data are expressed as mean ± standard deviation. The Student’s t-test was used to compare two groups for the results of the Western blot, CCK-8, cell migration, and cell invasion assays. P values of 0.05 or less were considered statistically significant.
Results
ATP6AP2 is Overexpressed in Breast Cancer
Based on the UALCAN data, ATP6AP2 mRNA expression was upregulated in breast cancer tissue (Figure 1A). The expression of ATP6AP2 in breast cancer samples at any age stages was higher than that in normal samples (Figure 1B). Patients in the age stage of 61–80 years owed the highest expression of ATP6AP2. ATP6AP2 was upregulated in samples with infiltrating ductal carcinoma (IDC), infiltrating lobular carcinoma (ILC) and mixed histology (Figure 1C). Whatever the status of menopause is, ATP6AP2 was higher in cancer samples than in normal samples (Figure 1D). Breast cancer with luminal type showed significantly higher expression of ATP6AP2 compared with other types of breast cancer (Figure 1E).
Validation of ATP6AP2 Expression in Breast Cancer by ONCOMINE Analysis
As ATP6AP2 was significantly overexpressed in ductal breast cancer through UALCAN analysis, we further validated ATP6AP2 mRNA expression levels in ductal breast cancer by using ONCOMINE database. ATP6AP2 was significantly overexpressed in breast cancer tissues in four datasets (Figure 2). In the Ma breast 4 dataset,15 ATP6AP2 was 2.061 fold elevated in breast cancer samples as compared with normal tissues (P = 6.31e-05). In the Radvanyi breast dataset,16 ATP6AP2 was 2.339 fold higher in breast cancer samples than in normal tissues (P = 0.003). In the Zhao breast dataset17 and Richardson breast 2 dataset,18 ATP6AP2 was also upregulated in ductal breast cancer samples with fold changes of 1.283 and 1.193, respectively.
 |
Figure 2 mRNA expression of ATP6AP2 in breast cancer (ONCOMINE). |
ATP6AP2 is Overexpressed in Luminal Type Breast Cancer Cell Lines
In GOBO analysis, ATP6AP2 expression was higher in luminal type breast cancer cell lines than in basal-A or basal-B type breast cancer cell lines (Figure 3A). Breast cancer cell lines with positive hormone receptor (HR) had the highest expression levels of ATP6AP2 (Figure 3B). ATP6AP2 expression in each cell lines is shown in Figure 3C.
ATP6AP2 Gene Alterations in Breast Cancer
Based on the cBioPortal Web resource online, the gene alterations of ATP6AP2 in breast cancer dataset (TCGA, PanCancer Atlas) are visualized in Figure 4A. High mRNA of ATP6AP2 was the most alterations in IDC and ILC, while deep deletion of ATP6AP2 was the least alterations. As Figure 4B showed, there were more ATP6AP2 diploid and less ATP6AP2 homozygous deletions.
ATP6AP2 Promotes MCF-7 Breast Cancer Cell Proliferation, Invasion and Migration
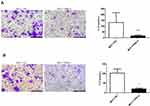
We used the MCF-7 luminal type breast cancer cell lines for next experiments. ATP6AP2 knockdown could significantly reduce ATP6AP2 protein (Figure 5A and B) and mRNA levels (Figure 5C) in MCF-7 cells. We next examined the effect of ATP6AP2 depletion on MCF-7 cell proliferation with the CCK-8 assay, and found that the OD value of cells transfected with PRR-siRNA was lower than that of cells transfected with NC-siRNA at 48 and 72 h (P < 0.01; Figure 6). In transwell assays, cell invasion (P < 0.001; Figure 7A) and migration (P < 0.05; Figure 7B) were markedly reduced by ATP6AP2 siRNA transfection relative to control cells. These findings indicated that ATP6AP2 knockdown inhibited migration and invasion in MCF-7 cells.
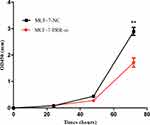 |
Figure 6 Effect of ATP6AP2 knockdown on MCF-7 cell proliferation. OD values resulting from the CCK-8 assay in MCF-7 cells. **P < 0.01. |
Functional Enrichment Analysis
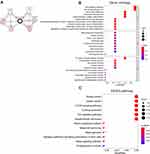
We utilized cBioPortal breast invasive carcinoma dataset (TCGA, PanCancer Atlas) to explore the biological interaction network of ATP6AP2 in breast cancer. By construction of network based on ATP6AP2 as the seed gene, we identified six genes connected with ATP6AP2 (Figure 8A). So, we performed functional enrichment analysis on these genes. CC of these genes was related to membrane. BP and MF of these genes were related to Wnt signaling (Figure 8B). The KEGG pathway analysis showed main enrichment in breast cancer, gastric cancer, mTOR signaling pathway, and Wnt signaling pathway (Figure 8C). The detailed GO and KEGG terms are presented in Figure 8B and C.
Discussion
In this study, we exploited data from public datasets and conducted in vitro experiments to explore the potential functions of ATP6AP2 in breast cancer. Our study revealed that ATP6AP2 was highly expressed in breast cancer tissues and inhibition of ATP6AP2 reduced breast cancer cell proliferation, invasion and migration.
The results of UALCAN and ONCOMINE analysis indicated that ATP6AP2 transcript level was significantly higher in breast cancer tissues than in normal tissues through. Specifically, ATP6AP2 was consistently upregulated in breast cancer regardless of age, histologic subtype and menopause status. Moreover, the highest expression level was observed in luminal type breast cancer, which was in accordance with the results of GOBO analysis. Thus, ATP6AP2 is potential diagnostic biomarker for breast cancer, although its clinical utility requires further clinical validation.
We analyzed ATP6AP2 gene alterations and copy number variations to explore the mechanism underlying the upregulation of ATP6AP2 expression in breast cancer. We found that mRNA upregulation was the major type of ATP6AP2 alteration and the most common type of copy number variations was diploid, which may account for the high expression of ATP6AP2 in breast cancer.
Our results also showed that ATP6AP2 can stimulate the proliferation of MCF-7 breast cancer cells. This is in line with previous study,8 in which ATP6AP2 promoted MCF-7 and SK-BR-3 breast cancer cell proliferation. In addition to this, we found that ATP6AP2 knockdown inhibited MCF-7 breast cancer cell invasion and migration ability, which indicated ATP6AP2 was capable of promoting breast cancer progression. These results suggest ATP6AP2 may serve as a potential therapeutic target for breast cancer and deserve further clinical research.
ATP6AP2, also known as PRR, is the member of rennin-angiotensin system (RAS).19 Binding to prorenin or renin, it activates RAS. Through this process, it regulates blood pressure and electrolyte balance.20 However, our knowledge about RAS was mostly limited to its roles in cardiovascular and renal diseases. Recently, emerging evidence has revealed that PRR has diverse functions in various cancers through the RAS.7,21,22 More importantly, all members of RAS including prorenin, renin, angiotensinogen, angiotensin I converting enzyme, and angiotensin receptors are aberrantly expressed in breast cancer tissues.23 Accordingly, ATP6AP2 may function through RAS to influence breast cancer development or progression.
Besides exerting RAS-related function, it can act as an adaptor protein between vacuolar-type ATPase and Wnt receptor as well.24,25 As a component of the Wnt receptor complex, ATP6AP2 is essential for Wnt/β-catenin signaling.25 ATP6AP2, as reported, is involved in ductal tumorigenesis in the pancreas through Wnt/β-catenin signaling. ATP6AP2 knockdown reduced pancreatic ductal adenocarcinoma cell proliferation and induced apoptosis of pancreatic ductal adenocarcinoma cell.26 In colorectal cancer, ATP6AP2 promoted colorectal cancer cells proliferation through Wnt/β-catenin signaling.27 In glioma, inhibiting ATP6AP2 also reduced the proliferative capacity of glioma cell lines by attenuating activation of Wnt/β-catenin signaling.6 In the present study, the results of functional enrichment analysis revealed that BP and MF of ATP6AP2 were related to Wnt signaling pathway and the most significant-enriched KEGG pathway was Wnt signaling pathway. These results support the findings of other studies, in which ATP6AP2 function through Wnt/β-catenin signaling.
Our study also identified several other important pathways associated with ATP6AP2 such as mTOR signaling pathway, Hippo signaling pathway and signaling pathways regulating pluripotency of stem cells. There are a number of evidence that these pathways are involved in multiple aspects of breast carcinogenesis including cell proliferation, adhesion, migration and invasion.28,29 As the versatile role of ATP6AP2, in-depth studies remain need to be conducted to explore the mechanism of ATP6AP2 in oncogenesis and progression of breast cancer.
Admittedly, our study has some limitations because we did not explore the real mechanism of ATP6AP2 in breast cancer. The roles of ATP6AP2 deserve further in vitro and in vivo studies because of its strong relevance with proliferation, invasion and migration of breast cancer.
Conclusion
ATP6AP2 is overexpressed in breast cancer. Moreover, ATP6AP2 could promote proliferation, invasion and migration of breast cancer cells.
Abbreviations
IARC, International Agency for Research on Cancer; ATP6AP2, adenosine triphosphatase H+ transporting accessory protein 2; PRR, (pro)renin receptor; qRT-PCR, quantitative real-time PCR; OD, optical density; CCK8, Cell Counting Kit-8 assay; cBioportal, cBio Cancer Genomics Portal; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular components; MF, molecular functions; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma; HR, hormone receptor.
Data Sharing Statement
All data used to support the findings of this article are included within this article and Supplemental materials information file.
Funding
This study was supported by Natural Science Foundation of Guangdong Province (2018A030313518).
Disclosure
Kankan Zhao and Mengchuan Wang are co-first authors for this study. The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Kinouchi K, Ichihara A, Sano M, et al. The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ Res. 2010;107(1):30–34. doi:10.1161/CIRCRESAHA.110.224667
3. Krop M, Lu X, Danser AH, Meima ME. The (pro)renin receptor. A decade of research: what have we learned? Pflugers Arch. 2013;465(1):87–97. doi:10.1007/s00424-012-1105-z
4. Delforce SJ, Lumbers ER, Corbisier DMC, et al. Expression of renin-angiotensin system (RAS) components in endometrial cancer. Endocr Connect. 2017;6(1):9–19. doi:10.1530/EC-16-0082
5. Beitia M, Solano-Iturri JD, Errarte P, et al. (Pro)renin receptor expression increases throughout the colorectal adenoma-adenocarcinoma sequence and it is associated with worse colorectal cancer prognosis. Cancers. 2019;11(6). doi:10.3390/cancers11060881
6. Kouchi M, Shibayama Y, Ogawa D, Miyake K, Nishiyama A, Tamiya T. (Pro)renin receptor is crucial for glioma development via the Wnt/beta-catenin signaling pathway. J Neurosurg. 2017;127(4):819–828. doi:10.3171/2016.9.JNS16431
7. Wang J, Nishiyama A, Matsuyama M, Wang Z, Yuan Y. The (pro)renin receptor: a novel biomarker and potential therapeutic target for various cancers. Cell Commun Signal. 2020;18(1):39.
8. Ohba K, Suzuki T, Nishiyama H, et al. Expression of (pro)renin receptor in breast cancers and its effect on cancercell proliferation. Biomed Res. 2014;35(2):117–126. doi:10.2220/biomedres.35.117
9. Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol. 2011;671:3–34.
10. Chandrashekar DS, Bashel B, Balasubramanya S, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
11. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/S1476-5586(04)80047-2
12. Fredlund E, Staaf J, Rantala JK, Kallioniemi O, Borg A, Ringnér M. The gene expression landscape of breast cancer is shaped by tumor protein p53 status and epithelial-mesenchymal transition. Breast Cancer Res. 2012;14(4):R113. doi:10.1186/bcr3236
13. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
14. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
15. Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11(1):R7. doi:10.1186/bcr2222
16. Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2005;102(31):11005–11010. doi:10.1073/pnas.0500904102
17. Zhao H, Langerød A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15(6):2523–2536. doi:10.1091/mbc.e03-11-0786
18. Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi:10.1016/j.ccr.2006.01.013
19. Nguyen G, Delarue F, Berrou J, Rondeau E, Sraer JD. Specific receptor binding of renin on human mesangial cells in culture increases plasminogen activator inhibitor-1 antigen. Kidney Int. 1996;50(6):1897–1903. doi:10.1038/ki.1996.511
20. Nguyen G, Delarue F, Burckle C, Bouzhir L, Giller T, Sraer JD. Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J Clin Invest. 2002;109(11):1417–1427. doi:10.1172/JCI0214276
21. George AJ, Thomas WG, Hannan RD. The renin-angiotensin system and cancer: old dog, new tricks. Nat Rev Cancer. 2010;10(11):745–759.
22. Zavadil J, Böttinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi:10.1038/sj.onc.1208927
23. Herr D, Rodewald M, Fraser HM, et al. Potential role of renin-angiotensin-system for tumor angiogenesis in receptor negative breast cancer. Gynecol Oncol. 2008;109(3):418–425. doi:10.1016/j.ygyno.2008.02.019
24. Jansen EJ, Martens GJ. Novel insights into V-ATPase functioning: distinct roles for its accessory subunits ATP6AP1/Ac45 and ATP6AP2/(pro) renin receptor. Curr Protein Pept Sci. 2012;13(2):124–133. doi:10.2174/138920312800493160
25. Cruciat CM, Ohkawara B, Acebron SP, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327(5964):459–463. doi:10.1126/science.1179802
26. Shibayama Y, Fujimori T, Nguyen G, et al. (Pro)renin receptor is crucial for Wnt/β-catenin-dependent genesis of pancreatic ductal adenocarcinoma. Sci Rep. 2015;5:8854. doi:10.1038/srep08854
27. Wang J, Shibayama Y, Zhang A, et al. (Pro)renin receptor promotes colorectal cancer through the Wnt/beta-catenin signalling pathway despite constitutive pathway component mutations. Br J Cancer. 2019;120(2):229–237. doi:10.1038/s41416-018-0350-0
28. Sharma VR, Gupta GK, Sharma AK, et al. PI3K/Akt/mTOR intracellular pathway and breast cancer: factors, mechanism and regulation. Curr Pharm Des. 2017;23(11):1633–1638.
29. Shi P, Feng J, Chen C. Hippo pathway in mammary gland development and breast cancer. Acta Biochim Biophys Sin (Shanghai). 2015;47(1):53–59. doi:10.1093/abbs/gmu114
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.