Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
Associations Between Single Nucleotide Polymorphisms of Hypoxia-Related Genes and Capsule Formation in Hepatocellular Carcinoma
Authors Chen S, Duan Y, Zhang Y, Cheng L, Cai L, Hou X, Li W
Received 28 April 2023
Accepted for publication 12 August 2023
Published 10 October 2023 Volume 2023:10 Pages 1785—1797
DOI https://doi.org/10.2147/JHC.S417830
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr David Gerber
Shanshan Chen,1,2,* Youjia Duan,2,* Yongchao Zhang,2 Long Cheng,2 Liang Cai,2 Xiaopu Hou,2 Wei Li1,2
1Cancer Center, Beijing Tongren Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Cancer Center, Beijing Ditan Hospital, Capital Medical University, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Li, Cancer Center, Beijing Tongren Hospital, Capital Medical University, 2 West Ring South Road, Daxing District, Beijing, 100176, People’s Republic of China, Tel +86 15811029005, Email [email protected]
Purpose: Tumor capsule is an independent prognostic factor for patients with hepatocellular carcinoma (HCC) and used increasingly to guide clinical decision-making. Considering the genetic complexity for capsule formation and its potential association with hypoxia, the significance of the polymorphisms of hypoxia-related genes in capsule formation and HCC prognosis remains to be elucidated.
Patients and Methods: Peripheral blood samples from HCC patients were collected in this study. Single nucleotide polymorphism (SNP) genotyping was conducted by the iPLEX chemistry on a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Sequenom, Inc.). The demographic and clinical data for the patients were obtained through medical chart review and/or consultation with the treating physicians. SPSS 25.0, R 4.1.1, and PLINK toolset were used to perform statistical analysis.
Results: A total of 183 patients were enrolled, including 88 patients assigned to the capsule group and 95 to the non-capsule group. SLC2A1 rs841858 T allele, SLC2A1 rs2297977 T allele, STAT1 rs1547550 C allele, and STAT1 rs34997637 G allele were associated with significantly increased risk of capsule formation. The genotypes of SLC2A1 rs841858, SLC2A1 rs2297977, STAT1 rs34997637, and STAT1 rs1914408 were significantly associated with the formation of HCC capsule. The polymorphisms of STAT1 rs2066802, STAT1 rs12693591, and HIF1A rs2057482 showed close relationship with the prognosis of HCC patients in the capsule group, while the genotype distributions of CTNNB1 rs4135385, IFNG rs1861494, and SERPINE1 rs2227631 were closely related to the survival of patients in the non-capsule group. Further haplotype analysis suggested that SLC2A1 block 1 and STAT1 block 2 were related to the susceptibility of HCC capsule.
Conclusion: The polymorphisms of the hypoxia-related genes (HIF1A, SERPINE1, IFNG, STAT1, CTNNB1, and SLC2A1) were correlated with the formation of HCC capsule. Several SNPs in these genes also showed association with HCC prognosis except SLC2A1. Further functional studies are warranted to explore the underlying mechanisms.
Keywords: single nucleotide polymorphism, hypoxia, capsule formation, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common tumors in the digestive system, accounting for 85–90% of primary liver cancers.1 The morbidity and mortality rates of HCC are increasing yearly.2 Although resection and ablation could achieve the effect of radical cure, since most HCC patients are diagnosed at an advanced stage, they usually have lost the radical opportunity with limited treatment options. In recent years, various advanced medical treatments have been applied in HCC management due to the development of science and medical technology. Currently, the commonly used local therapies for HCC include transarterial chemoembolization (TACE) and transarterial radioembolization (TARE).3,4 In terms of systematic therapy, various molecular-targeted drugs and immune-oncology drugs have been developed to improve the quality of life and prolong the survival time of patients.5,6 However, the progression and recurrence rates of HCC remain high, and growing attention has been paid to the mechanism of HCC development. It has been reported that various factors (eg, genetic susceptibility, immune response, and environmental factors) are involved in HCC carcinogenesis and progression.7,8 Tumor capsule, composed primarily of collagen and myofibroblast-like cells with elongated morphology and aSMA positivity in immunohistochemistry (IHC), is one of the pathological features of HCC.9,10 Capsule formation has been confirmed to be an independent risk factor for the prognosis of HCC patients.11,12 Ng IO et al reported that the disease-free and actuarial survival times of patients with encapsulated tumors were significantly better than those with nonencapsulated ones.12 Our previous study revealed a correlation between polymorphisms in several genes and the formation of tumor capsule.13
Mediated by hypoxia-inducible factor (HIF), hypoxia usually contributes to a series of changes in the microenvironment of HCC. As one of the most important components of the tumor microenvironment, hepatic stellate cells (HSCs) can be transformed into myofibroblast-like cells after activation, which are the most predominant cell types to form capsules.14 The production of extracellular matrix (ECM), especially collagen, is essential for fibrous capsule formation in tumor. Fibrocytes participate in tissue remodeling by producing ECM proteins and secreting various cytokines.15 It has been confirmed that the HIF-1α/LOX pathway is involved in remodeling the ECM.16 In addition, several enzymes involved in capsule formation and ECM remodeling, including prolyl hydroxylase, lysyl oxidase (LOX) family of secreted enzymes, and matrix metalloproteinases (MMPs), are regulated by HIF.17
Hypoxia-inducible factor 1 subunit alpha (HIF1A) encodes HIF-1α, which is the master regulator of hypoxic response and known to activate diverse signaling pathways in TME. One of its key downstream genes is serine protease inhibitor clade E member 1 (SERPINE1), encoding plasminogen activator inhibitor-1 (PAI-1) and acting on ECM. A recent study revealed that the stimulation with cancer-associated fibroblasts (CAFs) could promote the upregulation of PAI-1 in tumor-associated macrophages (TAMs) and enhance the malignant behavior of the HCC cells.18 Interferon gamma (IFNG) encodes IFN-γ, contributing to tumor immunity. A study confirmed that hypoxic conditions could lead to the downregulation of IFN-γ gene derived from CD8+ T lymphocytes, indicating a potential impact on tumor immune response by the hypoxic microenvironment.19 Our previous findings showed that IFN-γ was capable of inhibiting β-catenin signaling activity in HCC cells and inducing apoptosis.20 Several studies have demonstrated that IFN-γ involves in matrix deposition and fibrosis adhesion through regulation of PAI-1.21,22 Signal transducer and activator of transcription 1 (STAT1) is a key component of IFN-γ responses and has been considered to promote tumor development by enhancing tissue injury, remodeling, fibrosis, and inflammation.23 There is a complicated crosstalk between hypoxia signaling and Wnt/β-catenin pathway.24 β-catenin is encoded by catenin beta 1 (CTNNB1). Given its role in hepatic stellate cell biology and hepatic fibrosis development, mutations in CTNNB1 require due attention.25 Dickkopf-1 (DKK1) is a negative regulator in Wnt/β-catenin signaling pathway. Blocking the canonical Wnt pathway with DKK1 could affect HSC differentiation and liver fibrosis.26 The gene solute carrier family 2 member 1 (SLC2A1) codes for glucose transporter 1 (GLUT1) and participate in the basal uptake and storage of glucose in most cells. The transcription of the GLUT1 gene can be enhanced during prolonged exposure to hypoxia.27 GLUT1 expression has shown tight association with MMP-2 expression and cell invasiveness in human cancer cell lines, suggesting a potential role in capsule formation.28,29
Tumor capsule has been shown as a prognostic factor for HCC patients and used increasingly to guide clinical decision-making.30,31 The above mentioned studies suggest that hypoxia-related genes may play an important role in the formation of HCC capsule. In light of the genetic complexity of capsule formation and its potential association with hypoxia, the significance of the polymorphisms of hypoxia-related genes in capsule formation remains to be elucidated and needs further investigation. Therefore, we conducted this study to explore the potential correlation between single nucleotide polymorphisms (SNPs) in critical hypoxia-related genes and capsule formation.
Patients and Methods
Study Population
The study used peripheral blood samples and clinical data obtained from HCC patients treated at a major tertiary teaching hospital in Beijing from May 2019 to January 2020. All procedures were performed according to the Declaration of Helsinki with the approval of the Ethics Committee. The participants signed a written informed consent before inclusion in the study. All cases were newly diagnosed and confirmed by pathological or radiological examination as HCC. Patients were classified into two groups according to absence or presence of HCC capsule. Patients with a history of any other malignancy were excluded from the study.
Demographic and Clinical Data
The following information was collected through medical chart review and/or consultation with the treating physicians: age, gender, etiology, cirrhosis, HBV DNA, Child-Pugh classification, Barcelona Clinic Liver Cancer (BCLC) stage, tumor conditions (including tumor number, maximum tumor diameter, vascular invasion, and extrahepatic metastasis), laboratory tests (including alanine aminotransferase [ALT], aspartate aminotransferase [AST], total bilirubin [TBIL], albumin [ALB], and alpha fetoprotein [AFP]), and subsequent treatment.
Tag SNP Selection
Tag SNPs were selected based on dbSNP database (www.ncbi.nlm.nih.gov/SNP) and Ensembl website (http://grch37.ensembl.org/index.html). The SNP data of Chinese Han Beijing population were downloaded, and the SNP selection was performed using Haploview software. The cut-off of minor allele frequency (MAF) was set as >0.05, and linkage disequilibrium (LD) patterns with r2 were set as >0.8. In combination with the candidate SNPs previously reported, a total of 52 SNPs were finally selected in our study.
SNP Genotyping
The iPLEX chemistry on a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Sequenom, Inc.) was used for SNP genotyping. The genomic DNA extracted from peripheral blood samples was used as the template for polymerase chain reaction (PCR), and PCR assays were performed as previously described.13 Three primers were designed for each SNP, two for PCR amplification of approximately 120 bp product adjacent to the SNP site and one for single base extension. SNP genotypes were determined by analyzing primer extension products on mass spectrometry.
Statistical Analysis
SPSS 25.0, R 4.1.1, and PLINK toolset were used to perform statistical analysis. Hardy–Weinberg equilibrium (HWE) for each SNP in controls was evaluated using the chi-square test. Data were presented as mean ± standard deviation, median (interquartile range), or number (%) as appropriate. Student’s t-test was used to compare quantitative data, while Chi-square and Fisher’s exact tests were used for qualitative data. For non-normal continuous variables, Mann–Whitney U-test was conducted. Various genetic models (genotypic, dominant, and recessive) were estimated by PLINK toolset. Odds ratios (ORs) and 95% confidence intervals (CIs) were evaluated by logistic regression. Stratification analyses were further performed based on relevant clinical variables. Survival curves were obtained using the Kaplan–Meier method, and the survival outcomes were compared using the Log rank test. P < 0.05 was considered statistically significant. LD analyses and pairwise LD plots were generated using Haploview software.
Results
Patient Characteristics
A total of 183 patients were included in this study, of which 88 were assigned to the capsule group and 95 to the non-capsule group. The mean age for the study population was 59.10 ± 10.53 years. The baseline characteristics of the 183 patients are listed in Table 1. There were no significant differences between the two groups in demographics, tumor characteristics, laboratory tests, and treatment modalities.
 |
Table 1 Demographical Characteristics and Clinical Data of the Patients |
SNP Screening
Among the 52 SNPs selected to develop Fluidigm SNP assays, two SNPs (rs116908431 and rs12565724) failed genotyping. The genotype of rs1957757 was out of HWE in the control group (P < 0.01) and excluded from subsequent statistical analyses, as well as nine SNPs (rs1385129, rs77156365, rs1799889, rs118134071, rs2069717, rs2069709, rs2069707, rs41508050, and rs34005929) with MAF < 0.01. After SNP quality control, a total of 40 SNP loci were analyzed (Supplementary Table 1).
Individual SNP Association Analysis
The results of comparison of the allele distribution are shown in Supplementary Table 1. There were significant differences in the allele distribution of four SNPs between the two groups: SLC2A1 rs841858 (P = 0.015), SLC2A1 rs2297977 (P = 0.016), STAT1 rs1547550 (P = 0.042), and STAT1 rs34997637 (P = 0.016). Three genetic models (genotypic, dominant, and recessive) were used to compare the distribution of genotypes between groups (Supplementary Table 2). The genotype distribution of STAT1 rs1914408 was significantly different under the genotypic model (P = 0.048) and dominant model (P = 0.038). Significant difference was also observed in genotype distribution of STAT1 rs34997637 (P = 0.035) under the dominant model.
Shown in Figures 1 and 2 were the results of logistic regressions for the association with capsule formation. It was revealed that SLC2A1 rs841858 T allele (T/G, OR = 2.11, 95% CI: 1.14–3.91, P = 0.018), SLC2A1 rs2297977 T allele (T/G, OR = 1.80, 95% CI: 1.08–3.02, P = 0.025), STAT1 rs1547550 C allele (C/G, OR = 2.07, 95% CI: 1.01–4.26, P = 0.048), and STAT1 rs34997637 G allele (G/A, OR = 1.59, 95% CI: 1.07–2.37, P = 0.023) were significantly related to capsule formation (Figure 1). Compared with wildtype carriers, capsule formation was significantly associated with SLC2A1 rs841858 (GT/GG, OR = 2.15, 95% CI: 1.07–4.31, P = 0.032), SLC2A1 rs2297977 (GT/GG, OR = 2.23, 95% CI: 1.13–4.39, P = 0.020), STAT1 rs34997637 (GG/AA, OR = 2.53, 95% CI: 1.13–5.65, P = 0.023), and STAT1 rs1914408 (CT/CC, OR = 2.18, 95% CI: 1.15–4.11, P = 0.017) (Figure 2).
 |
Figure 1 Logistic regression analysis of associations between the alleles of selected SNPs and capsule formation. |
 |
Figure 2 Logistic regression analysis of associations between the genotypes of selected SNPs and capsule formation. |
We further explored the association between each SNP and capsule formation stratified by host characteristics (Supplementary Tables 3A–3I). The variant genotypes of SLC2A1 rs841858 GT, SLC2A1 rs2297977 GT, STAT1 rs34997637 GG, and STAT1 rs1914408 CT were consistently associated with a significantly increased odds of capsule formation in most subgroups. Notably, the variant genotypes of SERPINE1 rs2227667 GA and SERPINE1 rs2227692 TC were shown significant association with a decreased odds of capsule formation both in tumor diameter >5 cm subgroup and BCLC stage B subgroup. Furthermore, the TC genotype of HIF1A rs11549465 was significantly correlated with high capsule formation in subgroups of age ≤60 years, liver cirrhosis, and multinodular tumor, and a similar significant correlation was observed for the CT genotype of HIF1A rs2057482 in the subgroup of tumor diameter ≤5 cm. Other correlations with capsule formation reached significance including SLC2A1 rs841847 TC in multinodular tumor and bilobular involvement, STAT1 rs10173099 CC in male gender and absence of extrahepatic metastasis, STAT1 rs10208033 TC in male gender, solitary tumor, single lobar involvement, and absence of vascular invasion, STAT1 rs1547550 CG in absence of extrahepatic metastasis, STAT1 rs2280232 CA in presence of vascular invasion and BCLC C, and IFNG rs2430561 TA in bilobular involvement. Significant associations with absence of capsule were found with STAT1 rs3771300 GT in presence of extrahepatic metastasis, IFNG rs1861494 CC in age >60 years, CTNNB1 rs75288535 GA in female gender and BCLC C, and CTNNB1 rs4135385 AA in presence of extrahepatic metastasis, presence of vascular invasion, and BCLC C. The AG genotype of IFNG rs2069705 was significantly associated with capsule formation in bilobular involvement, while in single lobar involvement, it was significantly associated with absence of capsule.
Survival Analysis
The survival analysis showed that in capsule group, two variant genotypes (STAT1 rs2066802 GG and AG and STAT1 rs12693591 AA and CA) were risk factors for poor overall survival (OS) (median OS: 56.3 vs 108.6 months, P = 0.013; 44.7 vs 108.6 months, P = 0.001), while variant HIF1A rs2057482 TT and CT were protective factors (median OS: 108.6 vs 45.8 months, P = 0.004). In the non-capsule group, the homozygous variant genotypes CTNNB1 rs4135385 AA and IFNG rs1861494 CC were significantly associated with worse OS (median OS: 50.3 vs 96.8 months, P = 0.012; 45.6 vs 96.8 months, P = 0.022), while variant SERPINE1 rs2227631 AA and GA could prolong OS (median OS: 96.8 vs 50.3 months, P = 0.019) (Figure 3 and Supplementary Table 4).
Haplotype Block Structure and LD Analysis
Haplotype block structures were shown in Figures 4 and 5, Supplementary Figures 1–5. LD analysis was performed, and significant differences were indicated in several blocks between the cases and controls. Strong LD blocks were identified: block 1 (rs3864004 and rs75288535) and block 2 (rs1798802, rs11564465, rs4135385, and rs2293303) in CTNNB1; block 1 (rs2241529 and rs1569198) in DKK1; block 1 (rs1861494, rs2430561, and rs2069705) in IFNG; block 1 (rs2227667, rs2227692, and rs7242) in SERPINE1; block 1 (rs3754219, rs841853, and rs841847) in SLC2A1; block 1 (rs3771300, rs1914408, and rs2066804) and block 2 (rs34997637, rs12693591, rs10173099, rs2066802, rs10208033, and rs1467199) in STAT1. Among the blocks, there were significant differences in SLC2A1 block 1 (ACT, P = 0.047), STAT1 block 2 (GACGTC, P = 0.034), STAT1 block 2 (GCCATC, P = 0.013), and STAT1 block 2 (GATGCC, P = 0.028) between the cases and controls.
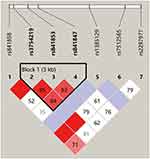 |
Figure 4 Haplotype block structure and linkage disequilibrium (LD) analysis of SLC2A1. |
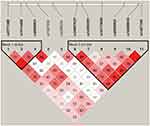 |
Figure 5 Haplotype block structure and linkage disequilibrium (LD) analysis of STAT1. |
Discussion
Tumor capsule is recognized as one of the pathological characteristics of HCC. Encapsulated HCC usually exhibits lower invasiveness due to the physical barrier effect of capsule. Capsule formation has been reported to be related to the developmental mode of HCC, the clinical manifestation of symptoms, and the prognosis of the disease.12 The presence of tumor capsule can often be detected by means of modern imaging techniques as well as ultrasound-guided or laparoscopy-guided needle biopsy of the liver. The mechanism of capsule formation is complex, involving multiple genes and signaling pathways. Given the important roles of hypoxia-related signaling pathways on ECM remodeling and tumor microenvironment, we have focused on the significance of SNPs of hypoxia-related genes in HCC capsule formation. HIF1A is one of the major regulators of hypoxic response, and SERPINE1, IFNG, STAT1, CTNNB1, and SLC2A1 are all critical downstream targets of HIF1A that could be involved in capsule formation. SERPINE1 encodes PAI-1 and affects ECM remodeling. IFNG has been reported to participate in the process of fibrosis and cooperate with activated STAT1 to function in tumor development. CTNNB1 and DKK1, important components of the Wnt pathway, deserve attention in HSC differentiation and fibrotic changes. SLC2A1 is closely related to MMP-2 expression and tumor invasion. Based on hypoxia-related signaling pathways, we selected the above seven genes (HIF1A, SERPINE1, IFNG, STAT1, CTNNB1, DKK1, and SLC2A1) and identified the association of the SNPs with the formation of HCC capsule.
HIF1A plays a central role in the formation of a hypoxic microenvironment within solid tumors. It can regulate multiple biological processes through the activation of numerous downstream target genes. The genetic association of HIF1A with cancer risk has been universally confirmed over the past few years.32,33 Guo et al have assessed the associations of three functional SNPs (rs2057482, rs1957757, and rs2301113) with clinicopathological parameters and prognosis of surgical HCC patients and found that HIF1A rs2057482 is significantly associated with clinical outcomes of Chinese HCC patients after surgery, especially in those with aggressive status.34 In addition, functional assay indicated the potential effect of rs2057482 on gene expression.34 In this study, HIF1A rs11549465 and HIF1A rs2057482 showed significant correlation with capsule formation in specific HCC patient subgroups, respectively. Further exploration from studies with larger sample size is required.
Cellular adaptation to hypoxia usually contributes to the transcriptional induction of a series of genes that participate in angiogenesis, glucose metabolism, and cell proliferation/survival.35 SLC2A1 which codes for GLUT1, is an important member involved in maintaining the growth and metabolism of cancer cells. Previous studies indicated that SLC2A1 (GLUT1) was highly expressed in a variety of cancer tissues and might be involved in the process of cancer development.36,37 The tumor metabolism status is determined by the expression of GLUT1 and ASCT2, and their metabolic index has also been identified as a promising prognostic predictor for HCC patients.38 Notably, SLC2A1 has a tight association with MMP2, a key gene for capsule formation. SLC2A1/MMP2 signaling pathway has been reported to be related to the invasiveness of various malignancies.28,29 Our findings suggested differences in genotype distribution of SLC2A1 rs841858 and SLC2A1 rs2297977, and this might provide the explanation for genetic variation of HCC capsule between the cases.
There is growing evidence that affirms the contribution of the pro-/anti-inflammatory cytokine balance and genetic factors to HCC and IFNG has been identified as such a susceptibility gene and risk factor.39,40 Although no study has yet investigated the association between IFNG gene polymorphisms and HCC capsules, IFN-γ, the protein encoded by IFNG, has been repeatedly confirmed to participate in the process of organ and tissue fibrosis.41,42 For example, the elevated levels of IFN-γ inhibit the expression of miR-351 in HSCs and interfere with hepatic fibrosis through activation of signal transducer and activator of transcription 1 and induction of IFN regulatory factor 2.41 Furthermore, IFN-γ pathway can play a role in cancer development through the activation of one of its major signal transducers STAT1.43 Significant association has been observed between polymorphisms in IFNG and STAT1 gene and HCC susceptibility.44,45 Our study suggested that STAT1 rs1547550, STAT1 rs34997637, and STAT1 rs1914408 were significantly associated with the formation of tumor capsule. In subgroup analyses, rs1861494, rs2430561, and rs2069705 of IFNG, and rs2280232, rs10208033, rs10173099, and rs3771300 of STAT1 also showed similar significance. Further research is needed to investigate the exact underlying mechanisms.
As a downstream target of hypoxia signaling pathways, SERPINE1 and its encoded protein PAI-1 mainly work in extracellular matrix and participate in the process of fibrin deposition and degradation. Mediated by plasminogen activation (PA) system, extracellular matrix remodeling is a critical step for changes in tumor biological characteristics. Therefore, SERPINE1 is probably involved in the formation of tumor capsule and affects the invasion and metastasis of HCC. Different from the other subgroups, the variant genotypes of SERPINE1 rs2227667 and SERPINE1 rs2227692 were shown significant association with capsule formation in tumor diameter >5 cm subgroup and BCLC B subgroup. We speculate that this may be correlated with the tumor status of patients. Cong et al found that changes of DNA stemlines and biological characteristics would occur along with the tumor size variations.46 In another study, more tumors were observed to be encapsulated in the larger group with diameter above 2 cm when compared to the smaller group with diameter below 2 cm.47 More researches should be performed to explore the effects of SNPs on capsule formation of certain HCC populations.
Wnt/β-catenin signaling pathway is well known to play an important role in cancer biology. In our previous studies, IFN-γ and β-catenin signaling have been proven to interrelate and influence each other in cancer cells.20,48,49 However, its crosstalk with hypoxia signaling pathway remains controversial. Genetic polymorphisms in CTNNB1 genes have been demonstrated to be closely associated with HCC risk and progression in a Chinese Han population.50 To our knowledge, this is the first study that has evaluated the association between polymorphisms in the Wnt/β-catenin pathway genes and HCC capsules. Significant results were observed in CTNNB1 rs75288535 (in subgroups of female and BCLC C) and CTNNB1 rs4135385 (in subgroups of presence of extrahepatic metastasis, presence of vascular invasion, and BCLC C). Further research on a larger sample size is needed.
Intratumoral hypoxia and genetic alterations can influence gene expression profiles, which have been associated with the prognosis of cancer patients.51,52 In our study, the genotypes of STAT1 rs2066802, STAT1 rs12693591, and HIF1A rs2057482 have shown prognostic value in the capsule group, and the genotypes of CTNNB1 rs4135385, IFNG rs1861494, SERPINE1 rs2227631 have shown prognostic value in the non-capsule group, suggesting that these hypoxia-related genes may be involved in the progression and prognosis of HCC patients with different phenotypes of tumor capsule. Encapsulation is able to limit tumor invasion to surrounding normal liver parenchyma, and patients with encapsulated tumors usually tend to have longer survival and lower recurrence rates after hepatectomy.53,54 In addition, tumor capsule can be used for evaluating the efficacy of TACE treatment for intermediate-stage HCC.31 For colorectal liver metastases, the good prognosis of encapsulated group seemed to be associated with reduced HIF-1α expression by the cancer.55 The related SNPs as prognostic markers for guiding medical decision-making deserved further investigation in future studies.
The proposed mechanism affecting capsule formation and HCC prognosis is shown in Figure 6 and the regulatory network is centered on HIF1A (HIF-1α). HIF1A (HIF-1α) could act on its targets SLC2A1 (GLUT1), IFNG (IFN-γ), STAT1, SERPINE1 (PAI-1), and CTNNB1 (β-catenin), respectively, and then engage on capsule formation. IFN-γ activates JAK1/2 and induces STAT1 phosphorylation. In addition, IFN-γ has crosstalks with the fibrinolytic system in which PAI-1 is a major component and the Wnt signaling where β-catenin plays key roles. STAT1 and PAI-1 interact and affect each other. All of them were proved to be associated with OS of HCC patients except SLC2A1 in this study. The precise mechanisms involved need to be verified through further studies.
 |
Figure 6 Proposed mechanism affecting capsule formation and HCC prognosis. One-way arrows represent unidirectional effects and two-way arrows represent bidirectional interactive relationships. |
Several limitations still existed in our research. Firstly, the relatively small sample size could have led to spurious association and low statistical power. More large prospective studies are warranted to identify genetic variants and confirm our findings. Secondly, some functional SNPs might be left out because our SNP selection was limited and did not cover all loci of the candidate genes, which might lead to low discriminatory power. Further future research should be more focused on the unknown SNP loci related to HCC capsule formation. Moreover, since our study was focused on association analysis instead of functional annotation, additional functional studies are needed to explore the underlying mechanism. At present, it is difficult to understand the actual functions of gene polymorphisms on HCC capsules and other biological behaviors without the exploration of the molecular mechanism. Despite these limitations, our work helps establish a foundation to better understand the association between the polymorphisms of hypoxia-related genes and HCC capsule, which is hopeful to provide more guidance for clinical assessment and decision-making.
Conclusion
Advancements in sequencing technologies and completion of the Human Genome Project have contributed to identifying genetic variation in human populations. SNP represents the most abundant class of genetic variations in the human genome and directly affects tumorigenesis and a series of malignant biological behaviors. In view of the genetic complexity of HCC capsule, a deep exploration of correlation between SNPs of hypoxia-related genes and capsule formation provides new references for guiding HCC prevention, diagnosis, and treatment in clinical practice. To conclude, our results demonstrated that the polymorphisms of hypoxia-related genes (HIF1A, SERPINE1, IFNG, STAT1, CTNNB1, and SLC2A1) were associated with the formation of HCC capsule, especially for SLC2A1 rs841858, SLC2A1 rs2297977, STAT1 rs1547550, STAT1 rs34997637, and STAT1 rs1914408. We have also identified several SNPs related to prognosis in capsule group and non-capsule group, respectively. These findings suggested that HIF1A-centric gene regulatory network might play a crucial role in the formation of HCC capsule, as well as the development of HCC. Our research provides new insights for analyzing HCC capsule, and future studies will be directed at increasing the sample size and mining more functional SNPs.
Abbreviations
HCC, hepatocellular carcinoma; IHC, immunohistochemistry; HIF, hypoxia-inducible factor; HSC, hepatic stellate cell; ECM, extracellular matrix; LOX, lysyl oxidase; MMP, matrix metalloproteinase; HIF1A, hypoxia-inducible factor 1 subunit alpha; SERPINE1, serine protease inhibitor clade E member 1; PAI-1, plasminogen activator inhibitor-1; CAF, cancer-associated fibroblast; TAM, tumor-associated macrophage; IFNG, interferon gamma; STAT1, signal transducer and activator of transcription 1; CTNNB1, catenin beta 1; DKK1, dickkopf-1; SLC2A1, solute carrier family 2 member 1; GLUT1, glucose transporter 1; SNP, single nucleotide polymorphism; BCLC, Barcelona Clinic Liver Cancer; ALT, alanine aminotransferase; AST, aspartate aminotransferase; TBIL, total bilirubin; ALB, albumin; AFP, alpha fetoprotein; MAF, minor allele frequency; LD, linkage disequilibrium; PCR, polymerase chain reaction; HWE, Hardy–Weinberg equilibrium; OR, odds ratio; CI, confidence interval; OS, overall survival; PA, plasminogen activation.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Beijing Ditan Hospital of Capital Medical University (KY2019-067) and was conducted according to the Declaration of Helsinki. All participants signed a written informed consent before inclusion in the study.
Author Contributions
All authors made significant contributions to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; agreed on the journal to which the article has been submitted; reviewed and agreed on all versions of the article before submission, during revision, the final version accepted for publication, and any significant changes introduced at the proofing stage; and agreed to take responsibility and be accountable for the contents of the article.
Funding
This work was supported by Beijing Municipal Science & Technology Commission No.Z221100007422026 and Beijing Talents Project.
Disclosure
The authors report no conflicts of interest in this work.
References
1. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi:10.1053/j.gastro.2007.04.061
2. Dasgupta P, Henshaw C, Youlden DR, Clark PJ, Aitken JF, Baade PD. Global trends in incidence rates of primary adult liver cancers: a systematic review and meta-analysis. Front Oncol. 2020;10:171. doi:10.3389/fonc.2020.00171
3. Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18(5):293–313. doi:10.1038/s41575-020-00395-0
4. Di Federico A, Rizzo A, Carloni R, et al. Atezolizumab-bevacizumab plus Y-90 TARE for the treatment of hepatocellular carcinoma: preclinical rationale and ongoing clinical trials. Expert Opin Investig Drugs. 2022;31(4):361–369. doi:10.1080/13543784.2022.2009455
5. Damaskos C, Garmpis N, Dimitroulis D, et al. Targeted therapies for hepatocellular carcinoma treatment: a new era ahead-a systematic review. Int J Mol Sci. 2022;23(22):14117. doi:10.3390/ijms232214117
6. Oura K, Morishita A, Tani J, Masaki T. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci. 2021;22(11):5801. doi:10.3390/ijms22115801
7. Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5(9):673–691. doi:10.1007/s13238-014-0065-9
8. Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99–111. doi:10.1016/j.ccr.2006.06.016
9. Ishizaki M, Ashida K, Higashi T, et al. The formation of capsule and septum in human hepatocellular carcinoma. Virchows Arch. 2001;438(6):574–580. doi:10.1007/s004280000391
10. Torimura T, Ueno T, Inuzuka S, Tanaka M, Abe H, Tanikawa K. Mechanism of fibrous capsule formation surrounding hepatocellular carcinoma. Immunohistochemical study. Arch Pathol Lab Med. 1991;115(4):365–371.
11. Fu S, Wei J, Zhang J, et al. Selection between liver resection versus transarterial chemoembolization in hepatocellular carcinoma: a multicenter study. Clin Transl Gastroenterol. 2019;10(8):e00070. doi:10.14309/ctg.0000000000000070
12. Ng IO, Lai EC, Ng MM, Fan ST. Tumor encapsulation in hepatocellular carcinoma. A pathologic study of 189 cases. Cancer. 1992;70(1):45–49. doi:10.1002/1097-0142(19920701)70:1<45::AID-CNCR2820700108>3.0.CO;2-7
13. Sun W, Zhang Y, Liu B, Duan Y, Li W, Chen J. Gene polymorphism of MUC15, MMP14, BRAF, and COL1A1 Is associated with capsule formation in hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2021;2021:9990305. doi:10.1155/2021/9990305
14. Barry AE, Baldeosingh R, Lamm R, et al. Hepatic stellate cells and hepatocarcinogenesis. Front Cell Dev Biol. 2020;8:709. doi:10.3389/fcell.2020.00709
15. Roife D, Fleming JB, Gomer RH. Fibrocytes in the tumor microenvironment. Adv Exp Med Biol. 2020;1224:79–85.
16. Tse AP, Sze KM, Shea QT, et al. Hepatitis transactivator protein X promotes extracellular matrix modification through HIF/LOX pathway in liver cancer. Oncogenesis. 2018;7(5):44. doi:10.1038/s41389-018-0052-8
17. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–439. doi:10.1038/nrc3726
18. Chen S, Morine Y, Tokuda K, et al. Cancer‑associated fibroblast‑induced M2‑polarized macrophages promote hepatocellular carcinoma progression via the plasminogen activator inhibitor‑1 pathway. Int J Oncol. 2021;59:2. doi:10.3892/ijo.2021.5239
19. De Ridder M, Jiang H, Van Esch G, et al. IFN-gamma+ CD8+ T lymphocytes: possible link between immune and radiation responses in tumor-relevant hypoxia. Int J Radiat Oncol Biol Phys. 2008;71(3):647–651. doi:10.1016/j.ijrobp.2008.03.014
20. Li W, Huang X, Tong H, et al. Comparison of the regulation of β-catenin signaling by type I, type II and type III interferons in hepatocellular carcinoma cells. PLoS One. 2012;7(10):e47040. doi:10.1371/journal.pone.0047040
21. Ohashi K, Yoshimoto T, Kosaka H, et al. Interferon γ and plasminogen activator inhibitor 1 regulate adhesion formation after partial hepatectomy. Br J Surg. 2014;101(4):398–407. doi:10.1002/bjs.9405
22. Kosaka H, Yoshimoto T, Yoshimoto T, Fujimoto J, Nakanishi K. Interferon-gamma is a therapeutic target molecule for prevention of postoperative adhesion formation. Nat Med. 2008;14(4):437–441. doi:10.1038/nm1733
23. Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97(6):439–447. doi:10.1111/j.1349-7006.2006.00197.x
24. Verras M, Papandreou I, Lim AL, Denko NC. Tumor hypoxia blocks Wnt processing and secretion through the induction of endoplasmic reticulum stress. Mol Cell Biol. 2008;28(23):7212–7224. doi:10.1128/MCB.00947-08
25. Monga SP. β-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology. 2015;148(7):1294–1310. doi:10.1053/j.gastro.2015.02.056
26. Miao CG, Yang YY, He X, et al. Wnt signaling in liver fibrosis: progress, challenges and potential directions. Biochimie. 2013;95(12):2326–2335. doi:10.1016/j.biochi.2013.09.003
27. Zhang JZ, Behrooz A, Ismail-Beigi F. Regulation of glucose transport by hypoxia. Am J Kidney Dis. 1999;34(1):189–202. doi:10.1016/S0272-6386(99)70131-9
28. Ito S, Fukusato T, Nemoto T, Sekihara H, Seyama Y, Kubota S. Coexpression of glucose transporter 1 and matrix metalloproteinase-2 in human cancers. J Natl Cancer Inst. 2002;94(14):1080–1091. doi:10.1093/jnci/94.14.1080
29. Ito H, Duxbury M, Zinner MJ, Ashley SW, Whang EE. Glucose transporter-1 gene expression is associated with pancreatic cancer invasiveness and MMP-2 activity. Surgery. 2004;136(3):548–556. doi:10.1016/j.surg.2004.05.032
30. Chao JS, Zhu Q, Chen DS, et al. Combined analysis of imaging tumor capsule with imaging tumor size guides the width of resection margin for solitary hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2022;21(6):551–558. doi:10.1016/j.hbpd.2021.12.009
31. Chen S, Peng Z, Zhang Y, et al. Lack of response to transarterial chemoembolization for intermediate-stage hepatocellular carcinoma: abandon or repeat? Radiology. 2021;298(3):680–692. doi:10.1148/radiol.2021202289
32. Wu LF, Xu GP, Zhao Q, Zhou LJ, Wang D, Chen WX. The association between hypoxia inducible factor 1 subunit alpha gene rs2057482 polymorphism and cancer risk: a meta-analysis. BMC Cancer. 2019;19(1):1123. doi:10.1186/s12885-019-6329-2
33. Li HN, He T, Zha YJ, et al. HIF-1α rs11549465 C>T polymorphism contributes to increased cancer susceptibility: evidence from 49 studies. J Cancer. 2019;10(24):5955–5963. doi:10.7150/jca.35716
34. Guo X, Li D, Chen Y, et al. SNP rs2057482 in HIF1A gene predicts clinical outcome of aggressive hepatocellular carcinoma patients after surgery. Sci Rep. 2015;5:11846. doi:10.1038/srep11846
35. Liu H, Wang Z, Yu S, Xu J. Proteasomal degradation of O-GlcNAc transferase elevates hypoxia-induced vascular endothelial inflammatory response†. Cardiovasc Res. 2014;103(1):131–139. doi:10.1093/cvr/cvu116
36. Liu XS, Gao Y, Wu LB, et al. Comprehensive analysis of GLUT1 immune infiltrates and ceRNA network in human esophageal carcinoma. Front Oncol. 2021;11:665388. doi:10.3389/fonc.2021.665388
37. Ancey PB, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. Febs j. 2018;285(16):2926–2943. doi:10.1111/febs.14577
38. Sun HW, Yu XJ, Wu WC, et al. GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PLoS One. 2016;11(12):e0168907. doi:10.1371/journal.pone.0168907
39. Zhou H, Wang L, Li X, et al. Interferon-γ +874A/T polymorphism and hepatocellular carcinoma risk: a meta-analysis. Med Sci Monit. 2015;21:689–693. doi:10.12659/MSM.892885
40. Kim HJ, Chung JH, Shin HP, et al. Polymorphisms of interferon gamma gene and risk of hepatocellular carcinoma in Korean patients with chronic hepatitis B viral infection. Hepato-gastroenterology. 2013;60(125):1117–1120. doi:10.5754/hge11333
41. He X, Sun Y, Lei N, et al. MicroRNA-351 promotes schistosomiasis-induced hepatic fibrosis by targeting the vitamin D receptor. Proc Natl Acad Sci U S A. 2018;115(1):180–185. doi:10.1073/pnas.1715965115
42. Attallah AM, El-Far M, Zahran F, et al. Interferon-gamma is associated with hepatic dysfunction in fibrosis, cirrhosis, and hepatocellular carcinoma. J Immunoassay Immunochem. 2016;37(6):597–610. doi:10.1080/15321819.2016.1179646
43. Yamashita N, Long M, Fushimi A, et al. MUC1-C integrates activation of the IFN-γ pathway with suppression of the tumor immune microenvironment in triple-negative breast cancer. J Immunother Cancer. 2021;9:1. doi:10.1136/jitc-2020-002115
44. Saxena R, Chawla YK, Verma I, Kaur JIFN. γ (+874) and not TNF-α (−308) is associated with HBV-HCC risk in India. Mol Cell Biochem. 2014;385(1–2):297–307. doi:10.1007/s11010-013-1838-9
45. Zhu ZZ, Di JZ, Gu WY, et al. Association of genetic polymorphisms in STAT1 gene with increased risk of hepatocellular carcinoma. Oncology. 2010;78(5–6):382–388. doi:10.1159/000320521
46. Cong WM, Wu MC. The biopathologic characteristics of DNA content of hepatocellular carcinomas. Cancer. 1990;66(3):498–501. doi:10.1002/1097-0142(19900801)66:3<498::AID-CNCR2820660316>3.0.CO;2-2
47. Wakasa K, Sakurai M, Okamura J, Kuroda C. Pathological study of small hepatocellular carcinoma: frequency of their invasion. Virchows Arch a Pathol Anat Histopathol. 1985;407(3):259–270. doi:10.1007/BF00710651
48. Li W, Tong H, Huang X, Wang W, Wu H, Lin S. High levels of β-catenin promote IFNγ-induced apoptosis in hepatocellular carcinoma cells. Oncol Lett. 2012;4(5):1092–1096. doi:10.3892/ol.2012.844
49. Bai M, Li W, Yu N, Zhang H, Long F, Zeng A. The crosstalk between β-catenin signaling and type I, type II and type III interferons in lung cancer cells. Am J Transl Res. 2017;9(6):2788–2797.
50. Li QM, Zhang FQ, Li YF, Xian QJ, Zhang YQ, Li P. Influence of polymorphisms in the Wnt/β-catenin pathway genes on hepatocellular carcinoma risk in a Chinese Han population. Medicine. 2017;96(12):e6127. doi:10.1097/MD.0000000000006127
51. Jing X, Yang F, Shao C, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18(1):157. doi:10.1186/s12943-019-1089-9
52. Luoto KR, Kumareswaran R, Bristow RG. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013;4(1):5. doi:10.1186/2041-9414-4-5
53. Chu KK, Chan SC, Fan ST, et al. Radiological prognosticators of hepatocellular carcinoma treated by hepatectomy. Hepatobiliary Pancreat Dis Int. 2012;11(6):612–617. doi:10.1016/S1499-3872(12)60232-X
54. Adachi E, Maeda T, Kajiyama K, et al. Factors correlated with portal venous invasion by hepatocellular carcinoma: univariate and multivariate analyses of 232 resected cases without preoperative treatments. Cancer. 1996;77(10):2022–2031. doi:10.1002/(SICI)1097-0142(19960515)77:10<2022::AID-CNCR9>3.0.CO;2-S
55. Siriwardana PN, Luong TV, Watkins J, et al. Biological and prognostic significance of the morphological types and vascular patterns in colorectal liver metastases (CRLM): looking beyond the tumor margin. Medicine. 2016;95(8):e2924. doi:10.1097/MD.0000000000002924
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

