Back to Journals » Infection and Drug Resistance » Volume 16
Association of Single Nucleotide Polymorphisms in Nucleotide-Binding Domain Leucine-Rich Repeat Protein 1 with Clostridioides difficile Colonization or Infection
Authors Tsai BY, Tsai PJ , Lee CC , Chiu CW, Lai YH, Lee JC, Ko WC , Hung YP
Received 6 October 2022
Accepted for publication 14 January 2023
Published 24 January 2023 Volume 2023:16 Pages 413—421
DOI https://doi.org/10.2147/IDR.S392510
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Bo-Yang Tsai,1,* Pei-Jane Tsai,1– 4,* Ching-Chi Lee,5,6 Chun-Wei Chiu,7 Yi-Hsin Lai,4 Jen-Chieh Lee,6 Wen-Chien Ko,6,8 Yuan-Pin Hung6– 9
1Institute of Basic Medical Sciences, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 2Department of Medical Laboratory Science and Biotechnology, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 3Department of Pathology, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 4Centers of Infectious Disease and Signaling Research, National Cheng Kung University, Tainan, Taiwan; 5Clinical Medicine Research Center, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 6Department of Internal Medicine, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 7Department of Internal Medicine, Tainan Hospital, Ministry of Health and Welfare, Tainan, Taiwan; 8Department of Medicine, College of Medicine, National Cheng Kung University, Tainan, Taiwan; 9Department of Microbiology & Immunology, College of Medicine, National Cheng Kung University, Tainan, Taiwan
*These authors contributed equally to this work
Correspondence: Yuan-Pin Hung; Wen-Chien Ko, Department of Internal Medicine, Tainan Hospital, Ministry of Health and Welfare, Tainan, Taiwan, Email [email protected]; [email protected]
Introduction: Nucleotide-binding domain leucine-rich repeat protein (NLRP) is critical in the inflammasome-activation pathway, which is important for host survival and the clearance of Clostridioides difficile. Therefore, the influence of NLRP1 polymorphisms on C. difficile colonization (CdC) or infection (CDI) was analyzed.
Materials and Methods: A prospective cohort study consisted of hospitalized adults was conducted from January 2011 to January 2013. Single nucleotide polymorphisms (SNPs) of NLRP1, including rs12150220, rs2670660, rs6502867, rs878329, rs8182352, rs3744717, and rs11078571, were incorporating in analyses. The episodes of CdC and CDI were the primary and secondary outcome, respectively.
Results: Of the total of 509 eligible patients, 376 (73.9%) had neither CdC nor CDI, 104 (21.8%) had CdC without developing CDI, and 29 (4.3%) developed CDI during the study period. Through multivariate analyses, comorbid diabetes mellitus (adjusted odds ratio [AOR] 1.59, P=0.04) and CC genotype in NLRP1 rs3744717 (AOR 1.70, P=0.02) were recognized as the risk factor of CdC. After adjusting the independent predictors of CDI, in terms of comorbid diabetes mellitus (AOR 3.18, P=0.005) and prior exposure to ceftazidime/ceftriaxone (AOR 2.87, P=0.04) or proton pump inhibitors (AOR 3.86, P=0.001), patients with CC+GC genotype in NLRP1, rs878329 (AOR 2.39, P=0.03) remained a higher risk of CDI.
Conclusion: For hospitalized adults, the association of CC genotype in NLRP1 rs3744717 and CdC as well as the CC+GC genotype in NLRP1 rs878329 and CDI was respectively evidenced. We believed the prompt identification of patients having specific genotype in NLRP1 would prevent and improve the quality of care in CDI.
Keywords: Clostridioides difficile infection, colonization, nucleotide-binding domain leucine-rich repeat protein 1, NLRP1, inflammasome, polymorphism
Introduction
Clostridioides difficile is one of the common etiologies of gastrointestinal infections, and can cause a wide spectrum of clinical manifestations, ranging from mild diarrhea or pseudomembranous colitis, to toxic megacolon, colon perforation, or death.1–8 C. difficile colonization (CdC), though asymptomatic, had been regarded as an important risk factor for C. difficile infection (CDI).1–8 The development of CDI is associated with several host characteristics, especially host innate immunity.1,9 Impaired host immunity, which occurs in conditions such as advanced age, the presence of comorbidities or polymorphisms of immune genes (such as IL-8 or toll-like receptor) is associated with an increased rate of C. difficile infection and disease recurrence.9,10 A critical property of the innate immune system is its ability to discriminate microbes from the “self” through the recognition of conserved microbial structures called pathogen-associated molecular patterns (PAMPs).11 The receptor-mediated mechanisms important for recognizing PAMP molecules involve Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). TLRs are membrane-bound proteins that sense PAMPs on the cell surface or in endosomes. Dissimilarly, NLRs recognize microbial molecules in the host cytosol.11
The human NLR family of cytoplasmic proteins contains 22 members, including 14 nucleotide-binding domain leucine-rich repeat proteins (NLRPs) and 5 NLRC, 1 NLRX, 1 neuronal apoptosis inhibitory protein (NAIP), and 1 class II trans-activator (CITTA) proteins.12 The members of the NLR family are essential during bacterial infection, especially those involving in the NLRP pathway,13 which is critical in the activation of inflammasomes that involve the participation of caspase-recruiting domain (ASC), caspase-1, and NLRP.14 Consequently, the activation of inflammasomes lead to enhanced IL-18 maturation and secretion.14
During CDI, C. difficile toxins A and B can lead to the release of IL-1 and to neutrophil recruitment via an inflammasome-activation pathway that is important for host survival and clearance of bacteria.15 Mice deficient in ASC, the adaptor protein in inflammasome assembly and activation, are highly susceptible to CDI.16 The role of the NLRP-inflammasome pathway in protective immunity against CDI was noted in a murine model.15,16 Among the NLR family members, the NLRP1, NLRP3, and pyrin pathways are critical for an effective immune response during bacterial infection.13,17 However, based on our published data, C. difficile induces inflammasome activation by neither the NLRP3 nor the pyrin inflammasome-dependent pathway.18 Because the role of NLRP1 in inflammasome activation during CDI is not clear and single nucleotide polymorphisms (SNPs) of NLRP1 genes reported to regulate inflammasome activation, cellular apoptosis, and the innate immune system among Asian populations,17,19 we hypothesized that the SNPs of NLRP1 vastly affect the occurrence of CdC or infection. Accordingly, a prospective cohort study was conducted to recognize the association between the SNPs of NLRP1 and CdC or CDI among hospitalized adults.
Materials and Methods
Study Design and Patient Selection
The prospective cohort study was conducted in the medical wards of Tainan Hospital, Ministry of Health and Welfare, a regional hospital in southern Taiwan, from January 2011 to January 2013. The study was approved by the institutional review board of National Cheng Kung University Hospital, Taiwan (approval number: B-ER-101-374), and written informed consent was obtained from all patients. The study was conducted in accordance with the Declaration of Helsinki. Partial information on this cohort has been published.20
Hospitalized patients aged at least 20 years were included, and within three months prior to the hospitalization, those experiencing CDI, receiving metronidazole or oral vancomycin, receiving a colectomy, having gastrointestinal infections due to identified pathogens, or lacking blood samples were excluded. The episodes of CdC and CDI were the primary and secondary outcomes, respectively. For capturing the outcome in each hospitalization, all the eligible patients had been followed-up from the first hospitalization (ie, the hospitalization with the patient inclusion) to the study endpoint (ie, the episode of CDI or June 2013).
Data Collection
Using a predetermined record form, medication information within three months prior to the first and index hospitalization (ie, the hospitalization with the first diagnosis of CdC or CDI), including antimicrobials, proton pump inhibitors, and H2-receptor antagonists, was queried. Clinical information during the initial index hospitalization, including patient demographics, nasogastric tube use, laboratory data, and underlying disease, was retrieved from medical records. When the patients were re-hospitalized, the observation timeframe was restarted from this readmission time, and stool was collected again.
The Diagnosis of CdC and CDI
Stool samples of the enrollees were collected within the first 48 hours of each hospitalization and every 7–10 days or at the onset of unexplained diarrhea during hospitalization. Immediately, the stool sample was delivered for C. difficile cultures within one hour after collection and plated on cycloserine-cefoxitin-fructose agar (CCFA) and cultured under anaerobic conditions. Multiplex polymerase chain reaction (PCR) was applied to detect tcdA, tcdB, and tcdC deletion in C. difficile isolates, as previously described.21
Diarrhea was defined as a change in bowel habit with three or more unformed stools per day for at least 2 days. Asymptomatic toxigenic CdC was defined as the detection of toxigenic C. difficile isolate from stool in the absence of diarrhea.22 A patient with tcdB-carrying C. difficile isolated from the feces in the presence of diarrhea without an alternative explanation was regarded to have CDI,22 whereas that with the lacking of diarrhea was regarded to have CdC. During the study follow-up, patients without the diagnosis of CdC and CDI were categorized as those with neither CdC nor CDI.
The Detection of SNPs in NLRP1 Genes
DNA extraction of blood samples was performed by the Genomic DNA Mini Kit (Geneaid Biotech, Taiwan) according to the manufacturer’s instructions. SNPs were detected by the ABI Prism® SNapShot™ Multiplex Kit and the predesigned TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA). SNPs of NLRP1 were selected from the SNP databank of National Center for Biotechnology Information (NCBI) for analysis according to two criteria. First, the SNPs had been reported to have a high frequency (minor allele frequency [MAF] of >0.05) in Asia in the GenBank® nucleic acid sequence database of the National Center for Biotechnology Information (NCBI).23 Second, the SNPs were associated with inflammatory or immunological responses. The SNPs of NLRP1 genes, including rs12150220, rs2670660, rs6502867, rs878329, rs8182352, rs3744717, and rs11078571, were included due to their linkage in regulating inflammasome activation, cellular apoptosis, and the innate immune system.17,19
Statistical Analysis
Statistical analysis was performed by statistical software (SPSS, version 13.0). Continuous data are expressed as the means ± standard deviations. The χ2 test or Fisher’s test was used for categorical variables, and the analysis of variance (ANOVA) or Student’s t test were used for continuous variables. The parameters with P values of less than 0.1 in the univariate analysis were put into the multivariate analysis with a hierarchical logistic regression model. A two-tailed P value of less than 0.05 was statistically significant.
Results
The Characteristics of the Cohort Patient
Among 554 hospitalized patients without a prior history of CdC or CDI, ten refused to participate in the study, and 35 had no available blood samples. Overall, 509 patients were enrolled (Figure 1). Of them, 281 (55.2%) were male, with an average age of 72.9 years, and 359 (70.5%) were nursing home residents. During the study period, 376 (73.9%) patients had neither CdC nor CDI, 111 (21.8%) had toxigenic CdC, and 22 (4.3%) without CdC experienced CDI. Of the 111 patients with toxigenic CdC, 7 subsequently suffered from CDI, and 104 remained in the status of CdC without evolution to CDI. Consequently, CDI was diagnosed in a total of 29 (5.7%) patients during the study follow-up. The crude mortality rates in patients with neither CdC nor CDI, those with CdC, and those with CDI were 14.9%, 15.5%, and 17.3%, respectively (P = 0.94). Their mean period of hospitalization was 30.0, 32.9, and 39.7 days, respectively (P = 0.63).
 |
Figure 1 The flowchart of patient selection. Abbreviations: CdC, Clostridioides difficile colonization; CDI, Clostridioides difficile infection. |
Clinical Information in the Non-CdC/CDI, CdC, and CDI Groups
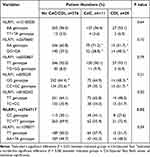
As shown in Table 1, the proportion of clinical variable between patients with neither CdC or CDI, those with CdC, and those with CDI was almost similar, in terms of patient demographics, comorbidities, and laboratory data. Patients with CDI or CdC were more likely to have comorbid diabetes mellitus than those with neither CdC nor CDI were. Compared to patients with neither CdC nor CDI, those with CDI or CdC were more likely to have the previous exposure of cephalosporins (particularly ceftazidime, ceftriaxone, or cefepime) and proton pump inhibitors (Table 2).
 |
Table 1 Comorbidities, Laboratory Data, and the tcdC Deletion in Adults with or Without Toxigenic Clostridioides Difficile Colonization (CdC) or Infection (CDI)* |
 |
Table 2 Previous Medications Within 3 Months Before Hospitalization in Adults with or Without Toxigenic Clostridioides Difficile Colonization (CdC) or Infection (CDI)* |
The SNPs of NLRP1 in the Non-CdC/CDI, CdC, and CDI Groups
In the SNP analysis of NLRP1, the prevalence rate of the AA genotype in rs12150220 was 96.1%, AA in rs2670660 was 64.4%, TT in rs6502867 was 91.6%, GG in rs878329 was 64.4%, TT in rs8182352 was 64.4%, and CC in rs3744717 was 34.0%. Patients with CdC exhibited the CC genotype in NLRP1 rs3744717 more often than did those with neither CdC nor CDI (Table 3). Additionally, a significant difference in the genotype of NLRP1 rs2670660 between CdC and CDI patients was disclosed, and a borderline difference in that of NLRP1 rs878329 between CDI patients and patients with neither CdC nor CDI was exhibited.
To overcome the interference of patient double-counting, patients with CdC or CDI were grouped together for the further analysis (Supplement Table 1). Similarly, only the CC genotype in NLRP1 rs3744717 remained more common among those with CdC or CDI than it was in those with neither CdC nor CDI (45.1% vs 30.1%, P = 0.002).
Independent Predictors of CdC or CDI
In the multivariate analysis, comorbid diabetes mellitus and the CC genotype in NLRP1 rs3744717 were two independent determinants of CdC (Table 4). When elucidating the patient characteristics of those with the CC genotype in NLRP1 rs3744717, they were observed to be older (75.5 ± 13.8 years vs 71.5 ± 15.7 years, P = 0.003) and more often female (52.6% vs 40.8%, P = 0.01) than those with the TC+TT genotype were (Supplement Table 2). However, the proportion of comorbidities was similar between the two groups.
 |
Table 4 Independent Variables of Developing Toxigenic Clostridioides Difficile Colonization (CdC) or Infection (CDI), Recognized by Multivariate Analyses* |
Through the multivariate analysis, comorbid diabetes mellitus, prior exposure to ceftazidime or ceftriaxone and proton pump inhibitors, and the CC+GC genotype in NLRP1 rs878329 were identified as independent risk factors of CDI (Table 4).
Discussion
The major finding in our study is that the presence of the CC genotype in NLRP1 rs3744717 is significantly associated with the development of CdC but not CDI, which is suggestive of the possible role of genetic background in the establishment of C. difficile intestinal colonization. In contrast, the prior receipt of certain medications (such as ceftazidime, ceftriaxone, or proton pump inhibitors) can predispose patients to the occurrence of CDI but not to CdC. The latter result is compatible with the observation that asymptomatic colonization with C. difficile is common among aged adults and that antibiotic therapy disrupts intestinal microbial composition and facilitates the development of CDI.24
The reported prevalence rate of each genotype among SNPs of NLRP1 in the Han population was 66.1% in the AA genotype of rs2670660,25 92.6–94.1% in the TT genotype of rs6502867,26 and 63.8–67.0% in the GG genotype of rs878329.26 These data were similar to the prevalence rates of SNP polymorphisms observed in our study. The polymorphism of NLRP rs3744717 has been proposed to be related to the increased activation of inflammasomes.17 Deficiency in inflammasome assembly and activation, which can lead to defects in neutrophil recruitment, has been shown to be associated with a higher susceptibility to CDI.16 Therefore, the result that the polymorphism of NLRP1 rs3744717 was associated with the acquisition of CdC that we observed in our study was reasonable.
Because of data from our published mouse study indicating that C. difficile induced inflammasome activation via neither the NLRP3 nor pyrin pathway,18 we focused on the potential effect of the NLRP1 family. In addition to NLRP1, the activation of NLRP3 is an alternative pathway for the formation of inflammasomes during CDI, as previously reported.15,16 In fact, among members of the NLR family, the NLRP1, NLRP3, and pyrin pathways are all critical for effective immune responses during bacterial infection.13,17 Therefore, NLRP1 and NLRP3 might both be important in activating the inflammasome pathway during CdC and CDI, but the role of NLRP3 polymorphisms remains to be explored.
The host variables disturbing gut microbiota and immune homeostasis and having been regarded as risk factors for CDI include antimicrobial exposure,27–32 advanced age,27–29 prior hospitalization,28 diabetes mellitus,33 and usage of proton pump inhibitors.29,34,35 Of note, patients with diabetes mellitus had higher rates of C. difficile colonization,36 severe CDI,33 and recurrent CDI.37 Furthermore, clinical therapy with several classes of antimicrobial agents, particularly third-generation cephalosporins,28,29 clindamycin,28,29,38 and fluoroquinolones,29,38 preceded the onset of CDI. Our study provides further clinical evidence that supports the presence of underlying metabolic diseases and prior usage of certain unfavorable drugs promote the consequences of CdC or CDI. Therefore, improvement of diabetic control and discontinuation of unnecessary antimicrobial agents are commonly regarded as indispensable measures in the treatment of patients with CDI.39
There were some limitations to the present work. First, only SNPs of NLRP1 were studied. The pathogenic role of polymorphisms of members of other NLR families, such as NLRP3, in the clinical contexts of colonic colonization or infection due to C. difficile remains undefined. Second, there were more than 100 cases of CdC, which outnumbered the number of CDI cases, and the NLRP1 rs3744717 polymorphism was significantly associated with CdC but not CDI. The small sample size of CDI patients may obscure our ability to investigate the impact of SNP of NLRP1. Third, we arbitrarily defined the presence of diarrhea as the indicator of CDI among those with CdC. Without further etiological investigations, multiple causes of diarrhea among the elderly render the overestimation of the prevalence of CDI possible. Fourth, although the enrollees were monitored for the appearance of CdC or CDI during hospitalization, the events occurring outside the hospital were missed. Fifth, the ribotypes or genetic classifications of C. difficile strains causing CDI or CdC were not examined. The variations in the pathogenic capability and temporal distribution of different ribotypes may lead to complex interactions between microbiological and host variables. Finally, the function of these NLRP SNPs was not analyzed, and the implications of these SNPs need further evaluations.
In conclusion, the association of SNP in NLRP1 and CdC or CDI has been discovered herein. However, with the acknowledgement of several limitations of the present study conducted among the Han population in a single center, the implications of the results should be extremely cautious before an external validation among non-Taiwanese populations is completed. In the future, the biological role of NLRP1 or other NLRP family in the clinical scenario of the occurrence of CdC or CDI events is warranted to be analyzed.
Data Sharing Statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Funding
The present study was supported by research grants from the Ministry of Science and Technology, Taiwan (109-2314-B-006-089-MY3, 110-2320-B-006-024 and 110-2314-B-675-001), Ministry of Health and Welfare, Taiwan (MOHW 110-TDU-B-211-124003), and National Cheng Kung University Hospital, Tainan, Taiwan (NCKUH-11004029).
Disclosure
All authors report no conflicts of interest relevant to this article.
References
1. Hung YP, Tsai CS, Tsai BY., et al. Clostridioides difficile infection in patients with hematological malignancy: a multicenter study in Taiwan. J Microbiol Immunol Infect. 2021;54(6):1101–1110. doi:10.1016/j.jmii.2021.02.002
2. Chiu CW, Tsai PJ, Lee CC, Ko WC, Hung YP. Inhibition of spores to prevent the recurrence of Clostridioides difficile infection - a possibility or an improbability? J Microbiol Immunol Infect. 2021;54(6):1011–1017. doi:10.1016/j.jmii.2021.06.002
3. Lee CC, Lee JC, Chiu CW, Tsai PJ, Ko WC, Hung YP. Clinical significance of toxigenic clostridioides difficile growth in stool cultures during the era of nonculture methods for the diagnosis of C. difficile infection. Microbiol Spectr. 2021;9(2):e0079921. doi:10.1128/Spectrum.00799-21
4. Tsai CS, Cheng YL, Chen JS, et al. Hypervirulent Clostridioides difficile RT078 lineage isolates from the river: a potential reservoir for environmental transmission. J Microbiol Immunol Infect. 2022;55(5):977–981. doi:10.1016/j.jmii.2022.05.002
5. Tsai BY, Chien CC, Huang SH, et al. The emergence of Clostridioides difficile PCR ribotype 127 at a hospital in northeastern Taiwan. J Microbiol Immunol Infect. 2022;55(5):896–909. doi:10.1016/j.jmii.2021.12.006
6. Zhang RF, Man YX, Bai YY, et al. Molecular characterization of Clostridioides difficile ribotype 027 in a major Chinese hospital. J Microbiol Immunol Infect. 2021;54(6):1179–1183. doi:10.1016/j.jmii.2021.01.003
7. Lee JC, Hung YP, Tsai BY, Tsai PJ, Ko WC. Severe Clostridium difficile infections in intensive care units: diverse clinical presentations. J Microbiol Immunol Infect. 2021;54(6):1111–1117. doi:10.1016/j.jmii.2020.07.012
8. Hung YP, Lee JC, Tsai BY, et al. Risk factors of Clostridium difficile-associated diarrhea in hospitalized adults: vary by hospitalized duration. J Microbiol Immunol Infect. 2021;54(2):276–283. doi:10.1016/j.jmii.2019.07.004
9. Lai YH, Tsai BY, Hsu CY, et al. The role of toll-like receptor-2 in Clostridioides difficile Infection: evidence from a mouse model and clinical patients. Front Immunol. 2021;12:691039. doi:10.3389/fimmu.2021.691039
10. DuPont HL, Garey K, Caeiro JP, Jiang ZD. New advances in Clostridium difficile infection: changing epidemiology, diagnosis, treatment and control. Curr Opin Infect Dis. 2008;21(5):500–507. doi:10.1097/QCO.0b013e32830f9397
11. Root-Bernstein R. Innate receptor activation patterns involving TLR and NLR synergisms in COVID-19, ALI/ARDS and sepsis cytokine storms: a review and model making novel predictions and therapeutic suggestions. Int J Mol Sci. 2021;22(4):2108. doi:10.3390/ijms22042108
12. Pedra JH, Cassel SL, Sutterwala FS. Sensing pathogens and danger signals by the inflammasome. Curr Opin Immunol. 2009;21(1):10–16. doi:10.1016/j.coi.2009.01.006
13. Jin C, Flavell RA. Molecular mechanism of NLRP3 inflammasome activation. J Clin Immunol. 2010;30(5):628–631. doi:10.1007/s10875-010-9440-3
14. Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys. 2019;670:4–14. doi:10.1016/j.abb.2019.02.008
15. Ng J, Hirota SA, Gross O, et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 2010;139(2):542–52, 52 e1–3. doi:10.1053/j.gastro.2010.04.005
16. Hasegawa M, Kamada N, Jiao Y, Liu MZ, Nunez G, Inohara N. Protective role of commensals against Clostridium difficile infection via an IL-1beta-mediated positive-feedback loop. J Immunol. 2012;189(6):3085–3091. doi:10.4049/jimmunol.1200821
17. Fernandes FP, Leal VNC, Souza de lima D, Reis EC, Pontillo A. Inflammasome genetics and complex diseases: a comprehensive review. Eur J Hum Genet. 2020;28(10):1307–1321. doi:10.1038/s41431-020-0631-y
18. Liu YH, Chang YC, Chen LK, et al. The ATP-P2X7 Signaling Axis Is an Essential Sentinel for Intracellular Clostridium difficile Pathogen-Induced Inflammasome Activation. Front Cell Infect Microbiol. 2018;8:84. doi:10.3389/fcimb.2018.00084
19. Li J, Yan M, Zhang Y, et al. Meta-analysis of the association between NLRP1 polymorphisms and the susceptibility to vitiligo and associated autoimmune diseases. Oncotarget. 2017;8(50):88179–88188. doi:10.18632/oncotarget.21165
20. Lai Y-H, Tsai B-Y, Hsu C-Y, et al. The role of toll-like receptor-2 in Clostridioides difficile infection: evidence from a mouse model and clinical patients. Front Immunol. 2021;12:548.
21. Hung YP, Cia CT, Tsai BY, et al. The first case of severe Clostridium difficile ribotype 027 infection in Taiwan. J Infect. 2015;70(1):98–101. doi:10.1016/j.jinf.2014.08.003
22. Loo VG, Bourgault AM, Poirier L, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365(18):1693–1703. doi:10.1056/NEJMoa1012413
23. Sayers EW, Bolton EE, Brister JR, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50(D1):D20–D6. doi:10.1093/nar/gkab1112
24. Schaffler H, Breitruck A. Clostridium difficile - from colonization to infection. Front Microbiol. 2018;9:646. doi:10.3389/fmicb.2018.00646
25. Sun X, Xia Y, Liu Y, et al. Polymorphisms in NLRP1 gene are associated with type 1 diabetes. J Diabetes Res. 2019;2019:7405120. doi:10.1155/2019/7405120
26. Sui J, Li H, Fang Y, et al. NLRP1 gene polymorphism influences gene transcription and is a risk factor for rheumatoid arthritis in han Chinese. Arthritis Rheum. 2012;64(3):647–654. doi:10.1002/art.33370
27. Dial S, Delaney JA, Barkun AN, Suissa S. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA. 2005;294(23):2989–2995. doi:10.1001/jama.294.23.2989
28. Vesteinsdottir I, Gudlaugsdottir S, Einarsdottir R, Kalaitzakis E, Sigurdardottir O, Bjornsson ES. Risk factors for Clostridium difficile toxin-positive diarrhea: a population-based prospective case-control study. Eur J Clin Microbiol Infect Dis. 2012;31(10):2601–2610. doi:10.1007/s10096-012-1603-0
29. Stevens V, Dumyati G, Fine LS, Fisher SG, van Wijngaarden E. Cumulative antibiotic exposures over time and the risk of Clostridium difficile infection. Clin Infect Dis. 2011;53(1):42–48. doi:10.1093/cid/cir301
30. Owens RC, Donskey CJ, Gaynes RP, Loo VG, Muto CA. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis. 2008;46(Suppl 1):S19–31. doi:10.1086/521859
31. Cho SM, Lee JJ, Yoon HJ. Clinical risk factors for Clostridium difficile-associated diseases. Braz J Infect Dis. 2012;16(3):256–261. doi:10.1590/S1413-86702012000300007
32. Dial S, Alrasadi K, Manoukian C, Huang A, Menzies D. Risk of Clostridium difficile diarrhea among hospital inpatients prescribed proton pump inhibitors: cohort and case-control studies. CMAJ. 2004;171(1):33–38. doi:10.1503/cmaj.1040876
33. Wenisch JM, Schmid D, Kuo HW, et al. Hospital-acquired Clostridium difficile infection: determinants for severe disease. Eur J Clin Microbiol Infect Dis. 2012;31(8):1923–1930. doi:10.1007/s10096-011-1522-5
34. Kwok CS, Arthur AK, Anibueze CI, Singh S, Cavallazzi R, Loke YK. Risk of Clostridium difficile infection with acid suppressing drugs and antibiotics: meta-analysis. Am J Gastroenterol. 2012;107(7):1011–1019. doi:10.1038/ajg.2012.108
35. Janarthanan S, Ditah I, Adler DG, Ehrinpreis MN. Clostridium difficile-associated diarrhea and proton pump inhibitor therapy: a meta-analysis. Am J Gastroenterol. 2012;107(7):1001–1010. doi:10.1038/ajg.2012.179
36. Lin HJ, Hung YP, Liu HC, et al. Risk factors for Clostridium difficile-associated diarrhea among hospitalized adults with fecal toxigenic C. difficile colonization. J Microbiol Immunol Infect. 2015;48(2):183–189. doi:10.1016/j.jmii.2013.08.003
37. Shakov R, Salazar RS, Kagunye SK, Baddoura WJ, DeBari VA. Diabetes mellitus as a risk factor for recurrence of Clostridium difficile infection in the acute care hospital setting. Am J Infect Control. 2011;39(3):194–198. doi:10.1016/j.ajic.2010.08.017
38. McFarland LV, Clarridge JE, Beneda HW, Raugi GJ. Fluoroquinolone use and risk factors for Clostridium difficile-associated disease within a Veterans Administration health care system. Clin Infect Dis. 2007;45(9):1141–1151. doi:10.1086/522187
39. Johnson S, Lavergne V, Skinner AM, et al. Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin Infect Dis. 2021;73(5):e1029–e44. doi:10.1093/cid/ciab549
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.