Back to Journals » OncoTargets and Therapy » Volume 10
Association of FHIT expression and FHIT gene hypermethylation with liver cancer risk: a PRISMA-compliant meta-analysis
Authors Zhang Y, Xu X, Chen Z, Zhao Z
Received 26 March 2017
Accepted for publication 17 May 2017
Published 20 June 2017 Volume 2017:10 Pages 3083—3093
DOI https://doi.org/10.2147/OTT.S138036
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Yaping Zhang,1 Xiao Xu,1 Zhiliang Chen,2 Zhenhua Zhao1
1Radiology Department, 2Department of Hepatobiliary Surgery, Shaoxing City People’s Hospital, Shaoxing, Zhejiang Province, People’s Republic of China
Background: There have been suggestions that fragile histidine triad protein (FHIT) expression and FHIT gene hypermethylation were crucial to the pathogenesis of liver cancer. However, the conclusions remained unclear because of small sample size, disease subtype, and different detection techniques. Therefore, we performed a meta-analysis to estimate the associations of FHIT expression and FHIT gene hypermethylation with liver cancer pathogenesis.
Methods: Studies that were published in electronic databases, such as PubMed, Web of Knowledge, China National Knowledge Infrastructure (CNKI), VIP, and WanFang, were retrieved and selected for the meta-analysis. Relative risk (RR) and 95% confidence interval (CI) were calculated to determine the correlations of FHIT expression and FHIT gene hypermethylation with liver cancer pathogenesis with Stata 12.0 software.
Results: A total of 17 papers that evaluated the associations of FHIT expression (14 articles) and FHIT gene methylation (3 articles) with liver cancer pathogenesis were included in this meta-analysis. In the overall analysis, the pooled relative risk was 1.93 (95% CI =1.72–2.17), which indicated a significant association between FHIT low expression and liver cancer risk. According to the results of clinical information, there were significant associations of FHIT expression with TNM-stage (RR =2.13, 95% CI =1.72–2.64), tumor size (RR =1.67, 95% CI =1.36–2.05), and merger of cirrhosis (RR =1.34, 95% CI =1.06–1.69) of liver cancer in the Chinese population. In addition, the FHIT gene hypermethylation was significantly associated with the risk of liver cancer (RR =1.45, 95% CI =1.08–1.93).
Conclusion: The FHIT expression and hypermethylation of FHIT gene were significantly associated with the risk of liver cancer, especially in the Chinese population. Furthermore, the results indicated significant associations between FHIT low expression and TNM-stage, tumor size, and merging of cirrhosis of liver cancer in the Chinese population.
Keywords: FHIT, expression, hypermethylation, liver neoplasms, meta-analysis
Introduction
Liver cancer, one of the most prevalent cancers, is the third leading cause of cancer-related deaths in the world.1 The incidence of liver cancer has reached one million worldwide every year and has a considerable geographic variation.2 The majority of liver cancer patients were found in developing countries such as China, Vietnam, North Korea, and Thailand.3 Although many therapeutic methods of hepatocellular carcinoma were developed, the prognosis was not satisfactory. A study reported that infection of the hepatitis B virus (HBV) and hepatitis C virus (HCV) was the main cause of the high prevalence of liver cancer in Asians.4 In addition, many risk factors, such as alcohol drinking, aflatoxin, obesity, diabetes, and tobacco use, and other genetic changes have been found to be associated with the development of liver cancer.5,6 Damage from liver cirrhosis was often seen during liver cancer. Excessive alcohol drinking was one of the common reasons of liver cirrhosis, which then led to the occurrence of liver cancer. If patients with liver cirrhosis were infected with HCV, they had a high risk of developing liver cancer. At the same time, aflatoxin B1 was another major risk factor for liver cancer due to the common food contamination in developing countries. If aflatoxin B1 was not metabolized completely, it could result in G-T transversions and ultimately induce the occurrence of liver cancer. In addition to these environmental factors, many genetic and epigenetic factors were involved in the initiation, progression, and metastasis of liver cancer.7,8 Epigenetic regulations, which consisted of altered gene expression without the change of DNA sequence, played an important role in the normal growth of cells, and its abnormality might lead to the occurrence of tumors.9 Therefore, a lot of epigenetic biomarkers, including the change of DNA methylation patterns, histone modifications, and the change of non-coding RNA expression profiles, were researched as the potential targets in the diagnosis, prognosis, and treatment of liver cancer.
FHIT, a member of evolutionary conserved histidine triad (HIT) proteins, belonged to oncosuppressor proteins which induced cell apoptosis. Studies have shown that transfection of FHIT cDNA affected the signal transduction of tumor cells and reduced the proliferation of cancer cell lines.10,11 The study of FHIT’s functions indicated that FHIT exerted its oncosuppressor activity by binding or mediation of fas, caspase-8, and PARP proteins.12 Deng et al reported that the overexpression of FHIT led to the activation of cell apoptosis factors.13 In recent years, several studies have been conducted to explore the association between the change of FHIT epigenetics and cancer risk. For instance, Lin et al found that the frequency of FHIT gene hypermethylation in myelodysplastic syndrome patients was higher than in normal persons.14 Moreover, it has been reported that the loss of FHIT was associated with the risk of gastric cancer, breast cancer, and lung cancer.15–17 And the aberrant methylation of FHIT gene had a significant association with the clinical stage and other clinical features.
Previously, many studies were performed to explore the role of FHIT expression and methylation in the development of liver tumor. The conclusions remained unsatisfactory because of small sample size. Therefore, in order to get more precise conclusions, we aimed to conduct a systematic meta-analysis to clarify the role of FHIT expression and FHIT gene methylation in the pathogenesis of liver cancer.
Materials and methods
Study strategy
A literature search was conducted using electronic databases such as PubMed, Web of Science, China National Knowledge Infrastructure (CNKI), VIP, and WanFang. The following terms were used to search the relevant articles: “Fragile histidine triad protein”, “FHIT”, “Hepatocellular carcinoma”, “HCC”, “liver cancer”, “liver tumor”, “liver carcinoma”, “methylation”, “hypermethylation”, “low expression”, and “expression”. MeSH terms and free-text words were used to search for eligible studies in the literature. The literature retrieval was updated on March, 2017. We checked the relevant reviews and references of included articles in detail and obtained the eligible literature. First, titles and abstracts of articles were read to initially confirm whether these articles were related to the associations of FHIT expression and FHIT gene methylation with liver cancer risk. Then, the full text of articles was scanned carefully and the relevant data were extracted.
Inclusion and exclusion criteria
The following inclusion criteria were applied to assess the relevant studies: 1) studies must have detailed diagnostic criteria of liver cancer; 2) studies must contain the data that evaluated the associations of FHIT expression and FHIT gene methylation with liver cancer; 3) studies were original research articles in English and Chinese language; and 4) if the data of the publications was the same, the latest study and more accurate data were included. Exclusion criteria were as follows: 1) reviews and meta-analysis were not included in this study; 2) studies that lacked sufficient data to estimate the relative risk (RR) and 95% CI; and 3) studies that contained duplicate data.
Data extraction and methodological assessment
The key components of eligible studies were recorded: publication year, the first author, studied population, type of cancer, cut-off value of immumohistochemical staining, number of patients, detection method of FHIT expression, sample type, frequency of FHIT gene methylation in case group and control group, and clinicopathological features. To acquire accurate data, two independent investigators were involved in the extraction of data. The methodological quality of all included studies was assessed using Newcastle-Ottawa Quality Assessment Scale (NOS) table. Studies that got a score of ≥6 were included in this meta-analysis.
Statistical analysis
RR and 95% CI were applied to estimate the associations of FHIT expression and FHIT gene hypermethylation with the clinical parameters of liver cancer. These clinical features included the TNM-stage, HBsAg, tumor size, liver cirrhosis, and liver capsule of liver cancer. Stata 12.0 software (Stata Corporation, College Station, TX, USA) was applied to calculate the RR and 95% CI. Forest plot and funnel plot were drawn to observe the heterogeneity and publication bias among studies. The chi-square based on Q-test and I2 test were performed to test the interstudy heterogeneity.18 Random effects model (DerSimonian and Laird method) and fixed effect model (Mantel-Haenszel method) were used to draw the forest plot and conduct the Q-test.19,20 If the absence of heterogeneity among studies was identified, fixed effect model was applied. Otherwise, the random effects model was used. The P-value and I2 statistic were used to quantitatively estimate the interstudy heterogeneity. The P-value of <0.05 or I2 statistic of >50% indicated a significant heterogeneity. Subgroup analysis and random effect model were performed if significant heterogeneity existed. Publication bias was evaluated by Begg’s rank correlation test and Egger’s weighted regression test, in which P<0.05 indicated a significant publication bias.21,22 Finally, sensitivity analysis was performed to assess the influence of a single study in the overall RR.
Results
Study characteristics
The process of literature search is summarized in Figure 1. A total of 110 articles were acquired in the initial retrieval by searching electronic databases. About 38 articles were excluded due to duplicate retrieval. Of the remaining 72 articles, 31 were obtained from Chinese databases and 41 publications from English databases. Through reading titles and abstracts, 44 irrelevant articles were excluded. Of the remaining 28 articles, 3 did not have the frequency of FHIT expression in case group and control group, 2 contained duplicate data, the samples of 4 were inappropriate, and 2 publications were about FHIT gene methylation and did not have sufficient frequency of FHIT gene methylation. Finally, 14 articles with 1,345 controls and 945 cases for FHIT expression were involved, while 3 articles with 75 controls and 124 cases for FHIT methylation were included..23–39 According to the extracted information of included studies, the frozen liver cancer tissues and normal or adjacent liver tissues were obtained through surgical operation to extract the DNA and detect the expression of FHIT and methylation of FHIT gene. The main characteristics of the eligible studies are shown in Table 1. The scores of methodological assessment of included studies ranged from 6 to 8.
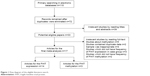  | Figure 1 Flow diagram of the eligible literature search. |
Association between FHIT expression and clinicopathological features of liver cancer
In total, the pooled results of the meta-analysis showed that persons with a low FHIT expression had a higher risk of liver cancer (RR =1.93, 95% CI =1.72–2.17). In addition, a significant association between FHIT low expression and liver cancer risk (RR =1.91, 95% CI =1.70–2.14) was found in Chinese population. The subgroup analysis based on sample type was conducted due to significant heterogeneity, and the results indicated a decreased heterogeneity and significant associations between FHIT low expression and liver cancer risk both in normal tissue (RR =2.00, 95% CI =1.72–2.32) and in adjacent tissue (RR =1.88, 95% CI =1.58–2.25). Moreover, there were significant associations of FHIT low expression with the TNM-stage (RR =2.13, 95% CI =1.72–2.64), tumor size (RR =1.67, 95% CI =1.36–2.05), and merger of cirrhosis (RR =1.34, 95% CI =1.06–1.69) with liver cancer. Interestingly, no significant association of FHIT low expression with the hepatitis B surface antigen (HBsAg) was found. According to the results of the TNM-stage, the frequency of FHIT low expression in III–IV stage patients of liver cancer was higher than that in the I–II stage patients of liver cancer. The expression of FHIT might be lower in the later-stage patients than that in the early-stage patients of liver tumor. Furthermore, the frequency of low FHIT expression was higher in the liver cancer tissue that merged with damage from liver cirrhosis. Random effect model was applied due to the existence of significant heterogeneity. No large heterogeneity was found other than the overall meta-analysis of liver cancer risk (Figures 2–5; Table 2).
  | Figure 3 Forest plot of the association between FHIT expression and TNM-stage of liver cancer. |
  | Figure 4 Forest plot of the association between FHIT expression and liver tumor size. |
Association between FHIT gene hypermethylation and liver cancer risk
Of all the 3 included studies with 75 controls and 124 cases, there was a significant association between FHIT gene hypermethylation and liver cancer risk (RR =1.45, 95% CI =1.08–1.93, P<0.05). No subgroup analysis based on ethnicity and disease type was conducted because little information was available. Furthermore, the results of Q test and the value of I2 revealed that no heterogeneity was detected in this meta-analysis. And the P-values of Begg’s test and Egger’s test were all >0.05, which suggested that the overall RR was stable (Figure 6).
  | Figure 6 Forest plot of the association between FHIT gene hypermethylation and liver cancer risk. |
Publication bias and sensitivity analysis
Begg’s test and Egger’s test were conducted and the results revealed that no significant publication bias was found in the meta-analysis of association of liver cancer pathogenesis with FHIT low expression. In the meta-analysis of FHIT expression, P<0.05 of Begg’s test indicated a significant publication bias. However, subgroup analysis based on sample type reduced the publication bias and nonsignificant value was found again. Sensitivity analysis was conducted by removing one study at a time. The pooled RR did not have significant changes, which indicated the stability of the result in this meta-analysis (Figures 7–8).
  | Figure 7 Funnel plot assessing the association of FHIT expression with liver cancer risk. |
  | Figure 8 Funnel plot of trim and filling method assessing the association of FHIT expression with liver cancer risk. |
Discussion
To the best of our knowledge, FHIT protein had a wide range of tumor-suppressive functions such as modulation of the DNA damage, downregulation of oncogene expression, modulation of tumor cells apoptosis, and the growth, cycle, invasion, and metastasis of cancer cells.40 FHIT could regulate the metabolism of Ap3A hydrolase and improve the activity of Ap3A which might restrain the growth of normal cells. Thus, the loss of FHIT protein expression might be one of the causes of unlimited reproduction in cancer cells.41 In most human normal tissues and cancer tissues, FHIT expression was detected and genetic alternations were often found. In some clinical studies, it has been reported that low FHIT expression had a significant association with the stage of many cancers. The previous study showed that the low expression of FHIT was associated with lymph node metastasis, lymphatic invasion, and the depth of invasion in gastric cancer, which suggested that FHIT expression might contribute significantly to the pathogenesis of gastric cancer.42 Moreover, some studies have reported that the change of FHIT low expression and FHIT gene hypermethylation had an important role to play in causing risk of lung cancer.43,44
In recent years, several studies have focused on the effect of genetic changes in FHIT protein on the development of hepatocellular carcinoma (HCC). Several studies have reported that the low expression of FHIT and FHIT gene mutations were shown in the HCC cell lines and primary HCC tissues.33,36,37 Aberrant FHIT transcripts were found in 39%–70% of HCC patients.45 Exon skipping, recognition of cryptic splice site, and use of alternate 5′ and 3′ splice sites led to the insertions of intronic sequences and further generated these aberrant transcripts.46 These transcripts affected the normal expression and function of FHIT protein. From previous studies, these alternations were often identified in the exon 5 and 6, and single-point mutations were very rare.37,46,47 In addition, some studies were also conducted to explore the change of FHIT protein expression other than these structural variations of FHIT gene. According to our literature search, most studies have focused on the FHIT expression and FHIT gene methylation in Chinese population. Therefore, a meta-analysis, assessing the associations of FHIT low expression and FHIT gene hypermethylation and liver cancer pathogenesis, was performed.
In the present study, 14 studies were included to investigate the correlation between FHIT expression and liver cancer risk and 3 studies were for FHIT gene methylation. Although a significant association of FHIT low expression with liver cancer risk was found, significant heterogeneity among studies was found. According to the results of subgroup analysis, the FHIT low expression resulted in a higher risk of liver cancer. At the same time, heterogeneity decreased a lot and no publication bias was found, which indicated a reliable result. In the included studies, Liu et al got an opposite result and other studies obtained the same result.27 Therefore, on the basis of the large sample size in this meta-analysis, we might conclude that there was a significant association between FHIT low expression and liver cancer risk. In the subgroup analysis based on clinical features in liver cancer, the pooled relative risk and 95% CIs suggested that there were significant associations of FHIT low expression with the TNM-stage, liver tumor size, and the merging of liver cirrhosis into liver cancer in Chinese population. Furthermore, in these included studies, Liu et al reported that the FHIT low expression had a higher frequency in the group of liver cirrhosis (patients who had liver cirrhosis and liver cancer).27 The results of Liu et al, Liang et al, Song et al, Nan et al, and Yuan et al indicated that the liver cancer patients of FHIT low expression often had a later stage of TNM.27,29,31,32,36 These results demonstrated that the expression of FHIT might be lower in patients with later-stage liver cancer. No significant publication bias and heterogeneity were found in the subgroup analysis based on sample type and clinical characteristics of liver cancer. Currently, complete liver capsule was very important in the diagnosis of liver cancer, which illustrated no infiltration of liver cancer cells. However, our data indicated that the status of FHIT expression was negatively associated with the integrity of the liver capsule. It is noteworthy that the FHIT low expression did not have a significant association with the existence of HBsAg. Thus, although almost 50% of HCC patients were affected with HBV infection, FHIT expression might not be associated with the infection of hepatitis B virus.47 In our meta-analysis, the sample of included studies mostly came from the hospital, which might result in some influences in the between-study heterogeneity. Furthermore, the frozen liver tumor tissues and control liver tissues were used to detect the status of FHIT expression and methylation of FHIT gene. According to the results of the clinical features in this meta-analysis, we could also analyze the FHIT expression and FHIT gene methylation of liver tumor tissues in different stages. Of importance, the association of clinical features with FHIT expression needed further investigation because of the single population, small sample size, and little clinical information.
Moreover, a significant association between FHIT gene hypermethylation and liver cancer risk was found based on a small sample size. On the basis of the results, no heterogeneity and publication bias among studies were found. However, although the results of the meta-analysis were significant and stable, the sample size was still small and little clinical information was available.
In addition, some limitations should be acknowledged in the present meta-analysis. First, cut-off values of immunohistochemistry were inconsistent, and this might reduce the accuracy of the results. Differences in evaluation criterion of staining, immunohistochemistry staining protocols, and cut-off values might lead to the heterogeneity among studies. Second, the studied population of this meta-analysis was mainly Chinese, and therefore, the results of this meta-analysis were mainly adapted to that population. The pathogenesis of liver cancer has a huge difference in different countries and regions. Hence, if we wanted to obtain more representative results all over the world, other studies in other populations and regions must be conducted. Third, the small number of included studies and the small sample size limited the statistical scope of the meta-analysis. Therefore, studies from a variety of countries and ethnicities should be conducted to improve the reliability of the conclusions.
Conclusion
In conclusion, FHIT low expression and FHIT gene hypermethylation were significantly associated with liver cancer risk, especially in Chinese population. The data also indicated that significant correlations of FHIT low expression with the TNM-stage, tumor size, and liver cirrhosis into liver cancer in Chinese population were found. Therefore, FHIT expression and FHIT gene hypermethylation might be good biomarkers for early diagnosis of liver cancer.
Acknowledgment
This work was supported by the Zhejiang provincial research plan for medical and health research (2013KYA205).
Disclosure
The authors report no conflicts of interest in this work.
References
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. | ||
Costanzo ES, Juckett MB, Coe CL. Biobehavioral influences on recovery following hematopoietic stem cell transplantation. Brain Behav Immun. 2013;30(Suppl):S68–S74. | ||
El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273. e1. | ||
Behnke M, Reimers M, Fisher R. The expression of embryonic liver development genes in hepatitis C induced cirrhosis and hepatocellular carcinoma. Cancers (Basel). 2012;4(3):945–968. | ||
Jain D, Nayak NC, Saigal S. Hepatocellular carcinoma in nonalcoholic fatty liver cirrhosis and alcoholic cirrhosis: risk factor analysis in liver transplant recipients. Eur J Gastroenterol Hepatol. 2012;24(7):840–848. | ||
Gassmann P, Spieker T, Haier J, Schmidt F, Mardin WA, Senninger N. Prognostic impact of underlying liver fibrosis and cirrhosis after curative resection of hepatocellular carcinoma. World J Surg. 2010;34(10):2442–2451. | ||
Verma M, Maruvada P, Srivastava S. Epigenetics and cancer. Crit Rev Clin Lab Sci. 2004;41(5–6):585–607. | ||
Verma M. Biomarkers for risk assessment in molecular epidemiology of cancer. Technol Cancer Res Treat. 2004;3(5):505–514. | ||
Calvisi DF, Simile MM, Ladu S, et al. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121(11):2410–2420. | ||
Siprashvili Z, Sozzi G, Barnes LD, et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci U S A. 1997;94(25):13771–13776. | ||
Ji L, Fang B, Yen N, Fong K, Minna JD, Roth JA. Induction of apoptosis and inhibition of tumorigenicity and tumor growth by adenovirus vector-mediated fragile histidine triad (FHIT) gene overexpression. Cancer Res. 1999;59(14):3333–3339. | ||
Roz L, Andriani F, Ferreira CG, Giaccone G, Sozzi G. The apoptotic pathway triggered by the Fhit protein in lung cancer cell lines is not affected by Bcl-2 or Bcl-x(L) overexpression. Oncogene. 2004;23(56):9102–9110. | ||
Deng WG, Nishizaki M, Fang B, Roth JA, Ji L. Induction of apoptosis by tumor suppressor FHIT via death receptor signaling pathway in human lung cancer cells. Biochem Biophys Res Commun. 2007;355(4):993–999. | ||
Lin J, Yao DM, Qian J, et al. Methylation status of fragile histidine triad (FHIT) gene and its clinical impact on prognosis of patients with myelodysplastic syndrome. Leuk Res. 2008;32(10):1541–1545. | ||
Geng X, Pu W, Tan Y, et al. Quantitative assessment of the diagnostic role of FHIT promoter methylation in non-small cell lung cancer. Oncotarget. 2017;8(4):6845–6856. | ||
Su Y, Wang X, Li J, Xu J, Xu L. The clinicopathological significance and drug target potential of FHIT in breast cancer, a meta-analysis and literature review. Drug Des Devel Ther. 2015;9:5439–5445. | ||
Wang HL, Zhou PY, Liu P, Zhang Y. Abnormal FHIT protein expression may be correlated with poor prognosis in gastric cancer: a meta-analysis. Tumour Biol. 2014;35(7):6815–6821. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ. 1998;316(7129):471. | ||
Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–680. | ||
Liu B, Wu XS, Ge JY, et al. Study of the expression differences of FHIT protein in 90 hepatocellular tissues with tissue chip method. J Kunming Med Univ. 2016;37(1):15–18. | ||
Lin J, Wang B, Hang AM, Wang XJ. The relationship between FHIT and WWOX expression and clinicopathological features in hepato cellular carcinoma. Fujian Med Univ. 2010;18:357–360. | ||
Che AW, Kang KF, Chen XW, Shi XC, Kang ZJ. Expressions of FHIT and Survivin in primary human hepatocellular carcinoma and study of relationship between them. Chin J Cancer Prev Treat. 2008;15:127–130. | ||
Fan ZJ, Zhao SK, Liu HR. The expressions and significance of FHIT, Survivin and Caspase-3 in hepatocellular carcinoma. Shaanxi Med J. 2008;37:1363–1366. | ||
Liu SR, Huang XH, Zou YH, Yang MJ, Liu ZM. Corresponding research of FHIT and AFP expressions of HCC with its clinicopathological grade and prognosis. J Clin Basic Res. 2008;15:1878–1881. | ||
Zhao P, Hou N, Lu Y. Fhit protein is preferentially expressed in the nucleus of monocyte-derived cells and its possible biological significance. Histol Histopathol. 2006;21(9):915–923. | ||
Liang FT, Zhang ZY, Liang FY. The role of FHIT expression in primary liver cancer. Infect Dis Info. 2005;18:68–69. | ||
Zheng J, Chen YL. The significance of FHIT expression in primary hepatocellular carcinoma. Pract Clin Med. 2005;6:44–45. | ||
Song WJ, Dou KF. Expression of FHIT and Ki- 67 in human hepatocellular carcinoma. Nw J Def med. 2005;26:129–131. | ||
Nan KJ, Ruan ZP, Jing Z, et al. Expression of fragile histidine triad in primary hepatocellular carcinoma and its relation with cell proliferation and apoptosis. World J Gastroenterol. 2005;11(2):228–231. | ||
Zekri AR, Bahnassy AA, Hafez M, et al. Alterations of the fragile histidine triad gene in hepatitis C virus-associated hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20(1):87–94. | ||
Xu RH, Yi JL. The expression of fragile histidine triad in primary hepatocellular carcinoma. J Clin Surg. 2004;12:412–414. | ||
Zhao P, Song X, Nin YY, Lu YL, Li XH. Loss of fragile histidine triad protein in human hepatocellular carcinoma. World J Gastroenterol. 2003;9(6):1216–1219. | ||
Yuan BZ, Keck-Waggoner C, Zimonjic DB, Thorgeirsson SS, Popescu NC. Alterations of the FHIT gene in human hepatocellular carcinoma. Cancer Res. 2000;60(4):1049–1053. | ||
Zekri AR, Bahnasy AA, Shoeab FE, et al. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J Adv Res. 2014;5(1):27–40. | ||
Zhang X, Li HM, Liu Z, et al. Loss of heterozygosity and methylation of multiple tumor suppressor genes on chromosome 3 in hepatocellular carcinoma. J Gastroenterol. 2013;48(1):132–143. | ||
Sun Y, Geng XP, Zu LX, et al. Clinicopathological significance of aberrant methylation of the fragile histidine triad gene in patients with hepatocellular carcinoma. Chin J Surg. 2006;44:609–612. | ||
Wali A. FHIT: doubts are clear now. ScientificWorldJournal. 2010;10:1142–1151. | ||
Zhao P, Liu W, Lu YL. Clinicopathological significance of FHIT protein expression in gastric adenocarcinoma patients. World J Gastroenterol. 2005;11(36):5735–5738. | ||
Nobili S, Bruno L, Landini I, et al. Genomic and genetic alterations influence the progression of gastric cancer. World J Gastroenterol. 2011;17(3):290–299. | ||
Tan S, Sun C, Wei X, et al. Quantitative assessment of lung cancer associated with genes methylation in the peripheral blood. Exp Lung Res. 2013;39(4–5):182–190. | ||
Verri C, Roz L, Conte D, et al. Fragile histidine triad gene inactivation in lung cancer: the European Early Lung Cancer project. Am J Respir Crit Care Med. 2009;179:396–401. | ||
Chen YJ, Chen PH, Chang JG. Aberrant FHIT transcripts in hepatocellular carcinomas. Br J Cancer. 1998;77:417–420. | ||
Gramantieri L, Chieco P, Di Tomaso M, et al. Aberrant fragile histidine triad gene transcripts in primary hepatocellular carcinoma and liver cirrhosis. Clin Cancer Res. 1999;5(11):3468–3475. | ||
Chang MH, Chen CJ, Lai MS, et al. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N Engl J Med. 1997;336(26):1855–1859. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




