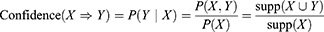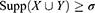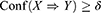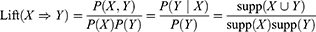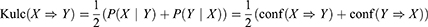Back to Journals » OncoTargets and Therapy » Volume 15
Association Mining Identifies MAL2 as a Novel Tumor Suppressor in Colorectal Cancer
Authors Wang K , Yang Y, Zheng S , Hu W
Received 6 April 2022
Accepted for publication 4 July 2022
Published 9 July 2022 Volume 2022:15 Pages 761—769
DOI https://doi.org/10.2147/OTT.S369670
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Kailai Wang,1,* Yanmei Yang,2,* Shu Zheng,1 Wangxiong Hu1
1Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310009, People’s Republic of China; 2Key Laboratory of Reproductive and Genetics, Ministry of Education, Women’s Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wangxiong Hu; Shu Zheng, Tel +86-571-87784606 ; +86-571-87784501, Fax +86-571-87214404, Email [email protected]; [email protected]
Introduction: Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide. However, the driver genes that promote CRC metastasis remain poorly understood. Association mining mines and extracts the repeated correlations and relevance in a dataset to predict the appearance of other data items according to the appearance of one item.
Methods: Here, the Apriori algorithm was used to find the frequent mutational gene sets (FMGSs) and hidden association rules (ARs) within these FMGSs from 383 CRCs with whole exome sequencing datasets. The weighted correlation network analysis (WGCNA) was used to identify the hub genes in CRC. CCK8, colony formation, cell migration and invasion assays were adopted to detect the roles of hub genes in CRC.
Results: Intriguingly, we found that MAL2 (myelin and lymphocyte protein 2) was associated with TP53 and APC in stage IV of CRC, and further subnetwork exploration based on WGCNA identified MAL2 as a potent hub gene. To validate the metastasis-related role of MAL2 in CRC, a lentivirus-based overexpression system was utilized to construct MAL2-overexpressing human CRC LOVO cells. Overexpression of MAL2 remarkably inhibited CRC cell proliferation and invasion.
Conclusion: Our results highlighted that MAL2 acts as a tumor suppressor in CRC and could serve as a potential therapeutic target.
Keywords: Apriori, CRC, frequent mutational gene sets, MAL2
Introduction
Colorectal cancer (CRC) is one of the most frequently occurring cancers and it ranks as the second leading cause of cancer-related mortality worldwide.1 In China, the incidence of CRC is on the rise, with 376,000 new cases and 191,000 deaths annually.2 The average five-year survival rate for CRC patients is only 65%, although the treatment of CRC is constantly improving.3 Metastasis is the major cause of CRC mortality.3,4 Although a compendium of mutational cancer driver genes capable of driving tumors has been identified,5 the driver genes, including both loss of function of tumor suppressors and gain of function of tumor promoters that favor CRC metastasis remain poorly understood.
Although some genes, such as KRAS, APC, and TP53, are frequently mutated in CRC, they are not metastasis-derived driver genes. Thus, we aimed to identify the specific driver mutation genes that are exclusively found in CRC stage IV by using the Apriori algorithm. Interestingly, we found that MAL2 (myelin and lymphocyte protein 2) was closely associated with TP53 and APC in CRC, but only in stage IV CRC. Further subnetwork exploration based on expression profiles found that MAL2 was a potent hub gene, with a high degree of gene connection in the gene coexpression network. It is tempting to believe that dysregulation of hub genes will substantially destabilize regulatory networks. Overexpression of MAL2 can significantly inhibit the proliferation, migration, and invasion of CRC cells, suggesting that MAL2 is a potential therapeutic target in CRC, especially for metastatic CRC (mCRC).
Materials and Methods
Identification of Frequent Mutational Gene Sets (FMGSs) and Subsequent Association Rule (AR) Mining in CRC
CRC somatic mutation data and clinical information were downloaded from the TCGA data portal (02/03/2015) and combined by their unique sample IDs. Silent mutations and any mutations in RNA, the 5’ untranslated region (UTR) and the 3’UTR were discarded. Retained mutation profiles were used to refine the mutated genes in 383 CRCs. Then, the Apriori algorithm was used to explore the FMGSs and ARs of mutated genes in the four stages of CRC. Frequent item sets (herein, FMGSs) are lists of items (herein, mutated genes) that commonly appear together, while ARs suggest that a strong relationship exists between two items. Briefly, let G = {g1, g2, …, gn} be a set of n genes called items. Let D = {t1, t2, …, tm} be a set of transactions (CRC patients) called the database. Each transaction in D has a unique patient ID and contains a subset of the genes in G. Starting by finding the frequent one-itemsets (k = 1), we iteratively generate candidate k+1 itemsets and check if they satisfy the support threshold. Note that the number of candidate itemsets decreases rapidly as k increases. A total of n+1 iterations are needed if the largest itemset has n items.
Once we find the frequent k-itemsets, we convert them into rules by splitting the k-itemsets (k ≥ 2) into antecedent (Genex, hereafter X) and consequent (Geney, hereafter Y). A rule is defined as an implication of the form X⇒ Y, where 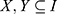 and
and 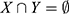 , meaning mutation of X probably leads to Y mutation. The itemsets X and Y are called antecedent (left-hand-side or LHS, one gene or more) and consequent (right-hand-side or RHS, one gene or more) of the rule. We started by putting a single gene in the consequent and k−1 genes in the antecedent. An interesting AR is a rule that surpasses a user-specified minimum support and minimum confidence threshold. Support (X) is defined as the proportion of patients in each tumor stage that contains the itemset and the confidence of a rule is defined as follows:
, meaning mutation of X probably leads to Y mutation. The itemsets X and Y are called antecedent (left-hand-side or LHS, one gene or more) and consequent (right-hand-side or RHS, one gene or more) of the rule. We started by putting a single gene in the consequent and k−1 genes in the antecedent. An interesting AR is a rule that surpasses a user-specified minimum support and minimum confidence threshold. Support (X) is defined as the proportion of patients in each tumor stage that contains the itemset and the confidence of a rule is defined as follows:
Therefore, an AR X⇒Y will satisfy:
where σ and δ are user-defined manually.
By default, to obtain reliable rules, minimum support (σ) was set at 0.1, and confidence (δ) was set at 0.9. Lower support or confidence can give rise to more FMGSs and rules but will also lead to spuriously significant findings. In the meantime, the confidence of a rule X⇒Y does not measure the actual strength of the correlation and implications between X and Y and it sometimes can be deceiving. One simple way to weigh the correlation of X and Y is lift:
In brief, the occurrence of Y is independent of the occurrence of X if P(X∪Y) = P(X)P;1 otherwise, Y and X are dependent and correlated as events. Lift values < 1 and > 1 indicate that the occurrence of X is negatively or positively correlated with the occurrence of Y, meaning that the occurrence of X likely leads to the absence or occurrence of Y, respectively.
Given that the prevalent mutational heterogeneity in cancer and lift can be easily influenced by the number of null-transactions (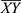 ), in combination with lift filtering (lift > 2), we used the Kulczynski measure6 for pattern exploration:
), in combination with lift filtering (lift > 2), we used the Kulczynski measure6 for pattern exploration:
Rules with Kulc > 0.7 were retained for further analysis.
Network Construction
All level three mRNA expression datasets (RNASeqV2) were obtained from TCGA (October 2018). Genes with an expression level < 1 (RSEM normalized counts) in more than 50% of the CRC samples were removed. Identification of differentially expressed genes (DEGs) was performed using the DEGSeq package for R/Bioconductor.7 Significant DEGs were selected according to a false discovery rate (FDR)-adjusted P value < 0.05 and fold change > 2 conditions. Then, the coexpression network was constructed by the weighted correlation network analysis (WGCNA) package for R8 and explored as in our previous work.9 The 50 top hub genes in the specific module were selected according to their connectivity degrees and visualized by VisANT.
Cell Culture
The human CRC cell line (LOVO) was purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). Cell lines were cultured in DMEM (HyClone) containing 10% (vol/vol) FBS (Biowest), 100 U/mL penicillin, and 100 mg/mL streptomycin and maintained at 37◦C in a humidified atmosphere of 5% CO2. A lentivirus-based MAL2 overexpression system was purchased from WZ Biosciences and transfected into the LOVO cells according to the manufacturer’s protocols.
Cell Proliferation Assays
We used a CCK-8 assay kit (CCK-8, Dojindo, Japan, CK04) to assess cell proliferation. Approximately 1000 LOVO cells were seeded in each well of 96-well plates. Subsequently, each day of the following five days, CCK-8 and complete medium (10:100) were mixed and added to each well. After incubating for 2 h at 37 °C away from light, the absorbance at 450 nm was used to measure the cell proliferation.
Cell Colony Formation Assay
Approximately 1000 cells were seeded in a 6-well plate containing complete culture medium and incubated in a 37 °C incubator. Colonies consisting of more than 50 cells after two weeks were counted. The assays were carried out in six replicates.
Cell Migration and Invasion Assay
Cell migration and invasion and siRNA knockdown assays were performed as described in our previous study.10 The sequences of MAL2 siRNA were as follows: sense: 5’-GGUCUGGCUUUACGAAGAUTT-3’ and antisense: 5’-AUCUUCGUAAAGCCAGACCTT-3’. The assays were carried out in triplicate.
Construction of Animal Models
The animal experiments were conducted according to the Animal Study Guidelines of Zhejiang University with approval number: 2018-286. Six four-week-old female nude mice (BALB/c, from the Zhejiang Chinese Medical University Laboratory Animal Research Center) were randomly divided into two groups: control (n = 3) and MAL2-OE (n = 3). The construction of the subcutaneous tumor xenograft mouse model has been described in our previous study.10
Statistical Analysis
All statistical analyses and graphical representations were performed in the R programming language (×64, version 3.5.1) and GraphPad Prism 7 unless otherwise specified.
Results
MAL2 Was Closely Associated with TP53 and APC Mutations in CRC
To identify novel metastasis-oriented driver genes in CRC, we first identified the differentially expressed genes (DEGs) in the four American Joint Committee on Cancer (AJCC) stages of CRC. However, no DEGs met the predefined statistical assumptions (fold-change > 2 and P < 0.05, nbinomTest) when advanced stage IV was compared with the other three stages. Thus, we speculated that driver mutation genes might play more important roles in CRC distant metastasis. To this end, we used the unsupervised learning method Apriori algorithm to explore the underlying relationship between two items under large somatic mutation data based on whole exome sequencing (WES). Intriguingly, the largest FMGS identified in CRC stage IV contained four genes (k = 4; ABCA7/TP53/APC/MAL2, LATS2/LURAP1L/TP53/APC, ABCA7/LURAP1L/TP53/APC, OBSCN/LURAP1L/TP53/APC, and TP53/APC/NEFH/KRAS), with a mutation frequency > 10% (support > 0.1). To further elucidate the putative correlation of these frequently mutated genes within FMGSs, the Apriori algorithm based on conditional probabilistic theory was used for mining ARs. Interestingly, we found that MAL2 mutation was closely associated with TP53 and APC in stage IV CRC (Table 1), indicating that MAL2 may act as an oncogene or tumor suppressor in CRC.
 |
Table 1 MAL2 Associated Rules Mined in Stage IV CRC |
MAL2 Acts as a Hub Gene in CRC
One way to decipher whether MAL2 acts as a driver gene in CRC is through the detection of weightiness in the regulatory network because hub genes may have a substantial impact on tumorigenesis and metastasis. Thus, WGCNA was adopted to explore the regulatory modules in CRC. Interestingly, we found six modules with different numbers of genes in CRC, and MAL2 acted as a hub gene in the green module (Figure 1), suggesting that it had a close association with CRC tumorigenesis if it was overexpressed or experience a loss of function.
MAL2 Regulates the Proliferation of CRC Cell Lines in vitro
It should be noted that an AR (X ⇒ Y) does not always uncover a causal relationship between them because there may be other hidden variables that cannot be deduced from the rule. To clarify the role of MAL2 in CRC tumorigenesis, a MAL2-overexpressing LOVO cell line (MAL2-ACT) was constructed (Figure 2A, t-test). A CCK-8 assay was performed to evaluate tumor cell proliferation. The results showed that the mean absorbance of the MAL2-ACT group was dramatically lower than that of the control group after Day 5 (P < 0.05, t-test, Figure 2B). Colony formation assays validated that increased expression of MAL2 could suppress LOVO cell proliferation, indicating that the upregulation of MAL2 impaired tumor cell growth (Figure 2C).
MAL2 Inhibits the Invasion of CRC Cell Lines in vitro
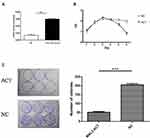
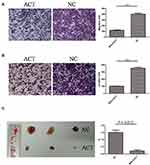
To determine whether MAL2 inhibits metastasis in CRC, a Transwell chamber with an 8-μm-pore filter membrane was used to measure tumor cell migration. Meanwhile, Matrigel-coated 8-μm-pore Transwell chambers were used to evaluate the cell invasion ability. Migrating/invading tumor cells on the underside of the filter membrane were stained and counted. The results revealed that both the migration and invasion abilities of MAL2-ACT cells were significantly weakened compared with those of control cells (P < 0.0001, t-test, Figure 3A and B). In contrast, both the migration and invasion properties of MAL2-KD LOVO cells were enhanced when MAL2-ACT cells was down-regulated by custom siRNAs (Figure S1).
MAL2 Suppresses CRC Tumorigenesis in vivo
To further confirm the tumor suppressor role of MAL2 in CRC, tumor xenograft mouse models were constructed. MAL2-ACT cells and controls (6×106 cells per mouse) were injected subcutaneously into mice. The mice were euthanized six weeks later, and the subcutaneous xenograft tumors were dissected and weighed (Figure 3C). The results showed that the average tumor weight of the MAL2-ACT group was significantly smaller than that of the control group (P = 0.013, t-test), suggesting that the increased expression of MAL2 remarkably inhibited CRC progression in vivo.
Discussion
It is known that distant metastasis is the major cause of cancer patient death, and continuous effort has been devoted to identifying driver genes of metastasis. In the present study, we found that MAL2 was a potential tumor suppressor that is closely related to APC and TP53 via Apriori association mining and validated this result by in vitro and in vivo experiments. As a member of the MAL family, MAL2 is a four-transmembrane protein whose gene is located on chromosome 8q24, commonly identified to be correlated with membrane apposition.11 Previous studies have reported that MAL2 functions in basolateral secretion, basolateral membrane protein delivery, and basolateral-to-apical transcytosis.12–14
MAL2 is recognized as an oncogene in some cancer types, such as breast cancer, ovarian carcinoma, and pancreatic ductal adenocarcinoma.15–18 However, Lopez-Coral et al19 recently found that MAL2 protein levels were decreased in malignant tissues compared to benign tissues in hepatocellular carcinoma (HCC), cholangiocarcinoma (CC), and renal cell carcinoma (RCC), and overexpression of MAL2 led to decreased cell migration, invasion and proliferation, which is consistent with our results (Figure 2). To reconcile the decreased MAL2 expression and its anti-oncogenic phenotype paradox, they proposed that enhanced MAL2 expression is associated with earlier stages of cancer progression and that its expression is repressed during the later stages and in metastases as Myc expression increases. This is somewhat similar to the findings that BRAF mutation can activate cancer cells in the early (neural crest phase) and middle (melanoblast phase) developmental stages, but it loses its the function of malignant transformation in mature melanocytes because of decreased expression of the chromatin-modifying enzyme ATAD2.20 Thus, oncogenic competence is mediated under certain cellular contexts (a combination of oncogenes, transcription factors, and developmentally regulated chromatin factors), which may reconcile the contradictory roles of MAL2 and allow it to have tumor-promoting and tumor-suppressing functions in different cancer types.
In conclusion, our results highlight MAL2 as a novel tumor suppressor and suggest that it may be a promising target for mCRC.
Data Sharing Statement
All datasets presented in this study are included in the article.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of the Second Affiliated Hospital, Zhejiang University School of Medicine.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81802883) and Fundamental Research Funds for the Central Universities (grant number 2018FZA7012) to Wangxiong Hu.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
4. Murphy CC, Harlan LC, Lund JL, Lynch CF, Geiger AM. Patterns of colorectal cancer care in the United States: 1990–2010. J Natl Cancer Inst. 2015;107(10). doi:10.1093/jnci/djv198
5. Martinez-Jimenez F, Muinos F, Sentis I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi:10.1038/s41568-020-0290-x
6. Uttayamakul S, Oudot-Mellakh T, Nakayama EE, et al. Genome-wide association study of HIV-related lipoatrophy in Thai patients: association of a DLGAP1 polymorphism with fat loss. AIDS Res Hum Retroviruses. 2015;31(8):792–796. doi:10.1089/aid.2014.0266
7. Wang L, Feng Z, Zhang X, Zhang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi:10.1093/bioinformatics/btp612
8. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi:10.1186/1471-2105-9-559
9. Hu W, Yang Y, Li X, et al. Multi-omics approach reveals distinct differences in left- and right-sided colon cancer. Mol Cancer Res. 2018;16(3):476–485. doi:10.1158/1541-7786.MCR-17-0483
10. Li X, Hu W, Zhou J, et al. CLCA1 suppresses colorectal cancer aggressiveness via inhibition of the Wnt/beta-catenin signaling pathway. Cell Commun Signal. 2017;15(1):38. doi:10.1186/s12964-017-0192-z
11. de Marco MC, Martin-Belmonte F, Kremer L, et al. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 2002;159(1):37–44. doi:10.1083/jcb.200206033
12. Marazuela M, Acevedo A, Garcia-Lopez MA, Adrados M, de Marco MC, Alonso MA. Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J Histochem Cytochem. 2004;52(2):243–252. doi:10.1177/002215540405200212
13. In JG, Striz AC, Bernad A, Tuma PL. Serine/threonine kinase 16 and MAL2 regulate constitutive secretion of soluble cargo in hepatic cells. Biochem J. 2014;463(2):201–213. doi:10.1042/BJ20140468
14. In JG, Tuma PL. MAL2 selectively regulates polymeric IgA receptor delivery from the Golgi to the plasma membrane in WIF-B cells. Traffic. 2010;11(8):1056–1066. doi:10.1111/j.1600-0854.2010.01074.x
15. Eguchi D, Ohuchida K, Kozono S, et al. MAL2 expression predicts distant metastasis and short survival in pancreatic cancer. Surgery. 2013;154(3):573–582. doi:10.1016/j.surg.2013.03.010
16. Byrne JA, Maleki S, Hardy JR, et al. MAL2 and tumor protein D52 (TPD52) are frequently overexpressed in ovarian carcinoma, but differentially associated with histological subtype and patient outcome. BMC Cancer. 2010;10:497. doi:10.1186/1471-2407-10-497
17. Bhandari A, Shen Y, Sindan N, et al. MAL2 promotes proliferation, migration, and invasion through regulating epithelial-mesenchymal transition in breast cancer cell lines. Biochem Biophys Res Commun. 2018;504(2):434–439. doi:10.1016/j.bbrc.2018.08.187
18. Fang Y, Wang L, Wan C, et al. MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J Clin Invest. 2021;131(1). doi:10.1172/JCI140837
19. Lopez-Coral A, Del Vecchio GJ, Chahine JJ, Kallakury BV, Tuma PL. MAL2-induced actin-based protrusion formation is anti-oncogenic in hepatocellular carcinoma. Cancers (Basel). 2020;12(2):422. doi:10.3390/cancers12020422
20. Baggiolini A, Callahan SJ, Trieu T, et al. Developmental chromatin programs determine oncogenic competence in melanoma. Science. 2021;373(6559). doi:10.1126/science.abc1048
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.