Back to Journals » International Journal of General Medicine » Volume 15
Association Between the Polymorphism of Steroid Hormone Metabolism Genes and High-Altitude Pulmonary Edema in the Chinese Han Population
Authors Gao H , Xu J, Ma Q, Tang F, Ga Q, Li Y, Guan W , Ge RL, Yang YZ
Received 21 October 2021
Accepted for publication 8 December 2021
Published 20 January 2022 Volume 2022:15 Pages 787—794
DOI https://doi.org/10.2147/IJGM.S345495
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Hui Gao,1,2 Jin Xu,2 Qiang Ma,2 Feng Tang,1 Qin Ga,1 Yuhong Li,3 Wei Guan,3 Ri-Li Ge,1 Ying-Zhong Yang1,2
1Research Center for High Altitude Medical Sciences, School of Medicine, Qinghai University, Qinghai, People’s Republic of China; 2Basic Medical Sciences, School of Medicine, Qinghai University, Qinghai, People’s Republic of China; 3Department of Respiration, Affiliated Hospital, Qinghai University, Qinghai, People’s Republic of China
Correspondence: Ying-Zhong Yang Tel/Fax +86 971 6143168
Email [email protected]
Purpose: Steroid hormone metabolism plays an essential role in high-altitude pulmonary edema (HAPE) progression. This study aimed to investigate the association between polymorphism in seven steroid hormone metabolism genes (STAR, HSD3B1, HSD3B2, CYP17A1, CYP21A2, CYP11B1, and CYP11B2) and HAPE susceptibility among Han Chinese.
Patients and Methods: A total of 41 tagSNPs in the seven genes were genotyped using Sequenom MassARRAY SNP assays from 169 HAPE patients (HAPE-p) and 309 matched Han Chinese individuals resistant to HAPE (HAPE-r). The genotypic and allele frequencies, odds ratios (ORs), and 95% confidence intervals (95% CIs) were calculated.
Results: Four SNPs, including the allele C of rs6203 (p = 0.034, OR [95% CI] = 1.344 [1.022− 1.767]) in HSD3B1, allele G of rs3740397 (p = 0.044, OR [95% CI] = 1.314 [1.007− 1.714]) and allele C of rs10786712 (p = 0.039, OR [95% CI] = 0.751 [0.572− 0.986]) in CYP17A1, and allele T of rs6402 (p = 0.006, OR [95% CI] = 0.504 [0.306− 0.830]) in CYP11B1, were significantly associated with HAPE. The distribution of the genotypes of these SNPs also significantly differed between the HAPE-p and HAPE-r groups. Moreover, six haplotypes (the linkage disequilibrium block including rs10883783, rs4919686, rs3740397, rs3824755, and rs10786712) of CYP17A1 were also significantly associated with HAPE.
Conclusion: The four SNPs located in HSD3B1 (rs6203), CYP17A1 (rs3740397 and rs10786712), and CYP11B1 (rs6402) and the six haplotypes of CYP17A1 are likely to have an effect on HAPE.
Keywords: steroid hormone metabolism gene, SNP, high-altitude pulmonary edema, susceptibility
Introduction
High-altitude pulmonary edema (HAPE) is a serious acute mountain sickness (AMS) that is triggered by the failure to acclimatize to high altitudes and hypoxia.1 The initial symptoms of HAPE include sympathetic overactivity, defective nitric oxide synthesis, exaggerated endothelin-1 synthesis, reduced fluid clearance from the alveolar space, and hypoxic pulmonary vasoconstriction, which can then develop into pulmonary hypertension, increased capillary pressure, and finally HAPE.2 Although hypoxia is a major trigger factor, the exact pathogenesis of HAPE remains unclear, and individuals usually differ in their susceptibility to HAPE at the same high altitude.3
At present, HAPE is considered a multifactorial disease involving both genetic and environmental risk factors.4,5 Many studies have shown that hypoxia-induced inflammation and steroid hormone metabolism at high altitudes may contribute to the development of HAPE.5–8 For example, some studies have reported that the salivary cortisol response after awakening differs between AMS+ and AMS- both at high and low altitudes, suggesting a link between hypothalamic-pituitary-adrenal (HPA)-axis homeostasis and AMS.9 In addition, high levels of plasma aldosterones have been detected in HAPE when compared with controls.10 Clinical observations have revealed that the onset time for HAPE is usually in the middle of the night, which is consistent with the period during which cortical hormone secretion is low. These studies suggest that an imbalance in endogenous cortical hormone secretion in HAPE patients may be a key underlying pathophysiological mechanism. The incidence of HAPE can be reduced using conventional drug-based treatments, such as acetazolamide and dexamethasone, which play an important role in the regulation of extracellular volume homeostasis and inflammatory reactions.2 This weight of evidence indicates that the adrenocortical hormone system and corresponding metabolism genes may significantly influence the pathophysiological HAPE process.
The biosynthesis of steroid hormones from cholesterol involves multiple enzymes, including cytochrome P450 enzymes (CYPs), hydroxysteroid dehydrogenases (HSDs), and steroid reductases. Studies have found that the polymorphisms of these enzymes are associated with a variety of diseases.11 Although many genetic association studies have identified dozens of genes associated with susceptibility to HAPE,12–19 few studies have reported a relationship between the genetic characteristics of steroid hormone metabolism genes and HAPE. Thus, in this study, we conducted a case–control association analysis to investigate the correlation between seven steroid hormone metabolism genes (STAR, HSD3B1, HSD3B2, CYP17A1, CYP21A2, CYP11B1, and CYP11B2) and HAPE in a Chinese Han population (Figure 1).
 |
Figure 1 Flowchart for this study. |
Materials and Methods
Subjects
In total, 169 Han Chinese who were admitted to Yushu & Hainan People’s Hospital between March 2010 and June 2020 due to the onset of HAPE 1−7 days after their arrival at the destination (>2500 m) were recruited by this study as HAPE patients (HAPE-p) and included in the final analysis. The diagnosis of HAPE was based on chest X-rays and standard diagnostic criteria.20 In total, 309 healthy controls that were resistant to HAPE (HAPE-r) were randomly selected from coworkers of the HAPE-p group to match the patients in terms of age, gender, ethnicity, and working conditions. These subjects remained healthy after working at the destination (>2500 m) for at least three months and did not suffer from HAPE or other high-altitude diseases. All subjects from both groups came from the same elevation and had the same ascent profile and working conditions. The study protocols were approved by the Ethics Committee of Qinghai University (Xining, China) and were conducted in accordance with the Declaration of Helsinki. All participants in this study signed informed consent forms.
Gene and SNP Selection and Genotyping Assays
Seven genes (STAR, HSD3B1, HSD3B2, CYP17A1, CYP21A2, CYP11B1, and CYP11B2) were selected because of their key roles in steroid hormone metabolism. The stellate protein encoded by the STAR gene is the first and rate-limiting step in cholesterol transport from the outer to the inner mitochondrial membrane.21 HSD3B (HSD3B1 and HSD3B2) is a key rate-limiting enzyme in the biosynthetic pathway for steroids, which produces aldosterone, estradiol, testosterone, and cortisol.11 The steroid 21 hydroxylase encoded by the CYP21 gene is a key enzyme in the synthesis of mineralocorticoid (aldosterone) and glucocorticoid (cortisol),22 while 17α-hydroxylase encoded by CYP17A is a key regulatory enzyme in the steroid hormone production pathway.23 CYP11B2 encodes aldosterone synthase, which catalyzes the final step in the biosynthesis of the major mineralocorticoid aldosterone, and 11β-hydroxylase, which is encoded by CYP11B1, is the final step in catalyzing the synthesis of cortisol.24
DNA was isolated from the whole blood cells of the participants with the use of a Gentra Puregene Blood Kit (Qiagen, 158389, Germany) following standard procedures. A total of 41 tagSNPs (a minor allele frequency >0.05, with an r2 threshold of 0.8) in the STAR, HSD3B1, HSD3B2, CYP17A1, CYP21A2, CYP11B1, and CYP11B2 genes have been identified in the HapMap Han Chinese population. The SNPs were genotyped using single-base extension detecting technology (iPLEX; Capital Bio Corporation, Beijing, China). The primers for PCR and single-base extension were designed using Sequenom MassARRAY Assay Design Genotyping Software and Tools (Sequenom, San Diego, CA, USA). PCR was conducted under the following thermal cycling conditions: 94°C for 4 min, followed by 94°C for 20 sec, 56°C for 30 sec, and 72°C for 1 min for 45 cycles, then 72°C for 4 min. PCR products were treated with shrimp alkaline phosphatase to remove free deoxyribonucleoside triphosphates, and the single-base extension reaction was performed in a system containing 2.0 mL of EXTEND MIX, 0.619 mL of ddH2O, 0.94 mL of Extend primer mix, 0.2 mL of iPLEX buffer plus, 0.2 mL of iPLEX terminator, and 0.041 mL of iPLEX enzyme (Sequenom, San Diego, CA, USA). The thermal cycling conditions were as follows: 94°C for 30 sec, followed by 94°C for 5 sec, 52°C for 5 sec, and 80°C for 5 sec for 40 cycles, then 72°C for 3 min. The purified extension products were dispensed onto a 384-element SpectroCHIP bioarray (Sequenom, San Diego, CA, USA), and mass spectrometric analysis was conducted using matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF; Sequenom, San Diego, CA, USA). The results were analyzed using TYPER 4.0 software (Sequenom, San Diego, CA, USA).
Statistical Analysis
SPSS software (version 17.0, SPSS, Inc., Chicago, USA) was used for the statistical analysis. Student’s t-tests and Fisher’s exact tests were used to assess the differences in the mean and frequency distributions of the demographic and clinical characteristics between the HAPE-p and HAPE-r groups, while the Hardy-Weinberg equilibrium (HWE) was used to assess the representativeness of the participants. Allele frequencies were calculated based on the genotype frequencies in the HAPE-p and HAPE-r groups, and the intergroup differences were estimated using χ2 tests. Haplotype frequencies and the expected number of haplotypes for each individual were calculated using SHEsis software (http://analysis.bio-x.cn).25 Differences were deemed significant at p < 0.05 for all comparisons.
Results
Basic Characteristics of the Study Population
A total of 169 patients with HAPE and 309 healthy controls were included in the study (Table 1). No significant differences were noted in the demographic characteristics (ie, gender or age) of the HAPE-r and HAPE-p groups, and no significant differences were observed in their clinical characteristics (ie, HR, SaO2, HGB, or Hct).
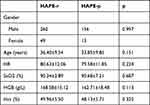 |
Table 1 Demographic and Clinical Characteristics of the Study Subjects |
Genotype and Allele Distribution
We calculated the genotypic distributions and allelic frequencies of 41 SNPs in the seven steroid hormone metabolism genes. The HWE was analyzed for the HAPE-p and HAPE-r groups using PLINK 2.0, and the 14 SNPs that did not conform to the HWE (corrected p < 0.05) were excluded (Table S1). The allele and genotype distributions of the remaining SNPs in the HAPE-p and HAPE-r groups are presented in Table S2. Of these, four were significantly associated with HAPE (Table 2). The minor allele C of rs6203 (p = 0.034, OR [95% CI] = 1.344 [1.022−1.767]) in HSD3B1 was significantly associated with the risk of HAPE. The distribution of the genotype between the HAPE-p and HAPE-r groups was also significantly different for this allele (p = 0.019, OR [95% CI] = 2.051 [1.117−3.767]). The minor allele G of rs3740397 (p = 0.044, OR [95% CI] = 1.314 [1.007−1.714]) and minor allele C of rs10786712 (p = 0.039, OR [95% CI] = 0.751 [0.572−0.986]) in CYP17A1 were significantly associated with HAPE. The distribution of the genotypes between the HAPE-p and HAPE-r groups was also significantly different for these two alleles (genotype GG of rs3740397: p = 0.049, OR [95% CI] = 1.694 [1.000−2.870]; genotype CC of rs10786712: p = 0.024, OR [95% CI] = 0.520 [0.293−0.923]). The minor allele T of rs6402 (p = 0.006, OR [95% CI] = 0.504 [0.306−0.830]) in CYP11B1 was significantly associated with a lower risk of HAPE. The distribution of the genotype between the HAPE-p and HAPE-r groups was also significantly different for this allele (p = 0.025, OR [95% CI] = 0.546 [0.320−0.932]).
 |
Table 2 Minor Allele and Genotype Frequencies for the SNPs Associated with HAPE in the Chinese Han Population |
Haplotype Analysis
As shown in Figure 2, rs10883783, rs4919686, rs3740397, rs3824755, and rs10786712 were present in a linkage disequilibrium (LD) block within CYP17A1. Haplotype analysis revealed that six haplotypes within that LD block differed significantly between the two groups (Table 3). Of these, the haplotypes TACCT (p = 0.049, OR [95% CI] = 1.536 [1.000−2.359]) and TACGT (p = 0.000, OR [95% CI] = 12.706 [5.078−31.795], p = 0.000) in CYP17A1 were associated with HAPE risk. In contrast, the haplotypes AAGGC (p = 0.038, OR [95% CI] = 0.492 [0.249−0.973]), TACCC (p = 0.039, OR [95% CI]= 0.551 [0.311−0.976]), TACGC (p = 0.000, OR [95% CI] = 0.014 [0.001−0.130]), and TAGGT (p = 0.025, OR [95% CI] = 0.720 [0.541−0.960]) in CYP17A1 were associated with a lower risk of HAPE.
 |
Table 3 Haplotype Effects for CYP17A1 (rs10883783-rs4919686-rs3740397-rs3824755-rs10786712) |
 |
Figure 2 Linkage disequilibrium for the five SNPs (rs10883783, rs4919686, rs3740397, rs3824755, and rs10786712) of CYP17A1 in the (A) HAPE-p group and (B) HAPE-r group. |
Discussion
To date, a number of genes associated with susceptibility to HAPE have been identified, including angiotensin-converting enzyme (ACE),12–14 endothelial nitric oxide synthase (eNOS),15,16 angiotensin (AGT) and its receptor (AGTR1),17 heat-shock protein 70 (HSP70),18 HLA,19 mitochondrial DNA (mtDNA),26 aldosterone synthase (CYP11B2),16 endothelin-1 (ET-1),12 pulmonary surfactant-associated protein A1 (SP-A1) and A2 (SP-A2),27 and β2 adrenergic receptor (ADRB2).28,29 However, few studies have investigated the association between the genetic characteristics of steroid hormone metabolism genes and HAPE. In this study, we conducted a case–control association analysis to investigate the correlation between seven steroid hormone metabolism genes and HAPE in a Chinese Han population. It was found that four SNPs located in HSD3B1, CYP17A1, and CYP11B1 were significantly associated with HAPE. Subsequent haplotype analysis revealed that six haplotypes in an LD block including five SNPs in CYP17A1 were significantly associated with HAPE.
The minor allele C of rs6203 (p = 0.034, OR [95% CI] = 1.344 [1.022−1.767]) in HSD3B1 was significantly associated with the risk of HAPE. HSD3B is a key rate-limiting enzyme in the steroid biosynthesis pathways that produce aldosterone, estradiol, testosterone, and cortisol. It has two tissue-specific isoforms (HSD3B1 and HSD3B2) that have different substrate affinities.30 In humans, a genetic variation in HSD3B1 can lead to an elevation in plasma aldosterone with a resultant increase in intravascular volume and hypertension.31 Research has found that rs6203 and rs1047303 in HSD3B1 are useful genetic markers for essential hypertension, while polymorphisms of HSD3B1 are associated with blood pressure and aldosterone levels.32
The minor allele G of rs3740397 (p = 0.044, OR [95% CI]= 1.314 [1.007−1.714]) and minor allele C of rs10786712 (p = 0.039, OR [95% CI] = 0.751 [0.572−0.986]) in CYP17A1 were significantly associated with HAPE, as were six haplotypes in CYP17A1 The microsomal enzyme cytochrome P450c17α, a key regulatory enzyme in the steroidogenic pathway, is encoded by CYP17A1, which is located on chromosome 10q24.3. It has two key roles in 17α-hydroxylase and 17, 20-lyase activity that determine the ability of adrenal glands and gonads to synthesize 17α-hydroxylated glucocorticoids (17α-hydroxylase activity) and/or sex steroids (17, 20-lyase activity).33 Various disease susceptibility variants of CYP17A1 have been widely reported. For example, genetic polymorphisms in CYP17A1 have been linked to variations in estrogen levels and to the increased risk of polycystic ovaries,34,35 breast cancer,36 and other estrogen-related diseases. CYP17A1 variants have also been reported to be associated with an increased risk of high-altitude polycythemia (HAPC) in Tibetans.37
The minor allele T of rs6402 (p = 0.006, OR [95% CI] = 0.504 [0.306−0.830]) in CYP11B1 was significantly associated with a lower risk of HAPE. CYP11B1 encodes 11β-hydroxylase, the enzyme that catalyzes the final step in the synthesis of cortisol.38 Some evidence suggests that the CYP11B1/B2 locus is important in hypertension and cardiovascular regulation.39 It has been reported that there is a significant association between polymorphisms in CYP11B2 and CYP11B1 and a genetic predisposition towards primary hyperaldosteronism.40
Several studies have reported that some genetic loci associated with lung-related diseases overlap with loci for pulmonary fibrosis.41–43 For example, two chronic obstructive pulmonary disease-associated loci (FAM13A and DSP) overlap with loci for lung function and pulmonary fibrosis,42 while five pulmonary fibrosis-associated loci (DPP9, DSP, FAM13A, IVD, and MUC5B) are also significantly associated with interstitial lung abnormalities.43 However, we did not find an explicit genetic association between HAPE and pulmonary fibrosis susceptibility in this study, and this has not been reported in other genetic analyses of HAPE. Of the seven selected genes, only CYP17A1 has been reported to be associated with HAPC in Tibetans on the Qinghai-Tibetan plateau.37 Thus, further research needs to be conducted to investigate the association between these genes and altitude sickness and/or other hypoxia-related diseases.
Another limitation of the student is that, due to the lack of large plateau populations, our analysis could only test a moderate number of subjects. We also did not attempt to employ population stratification in our sample, which is considered to be the most important confounder in genetic association mapping. Moreover, we did not measure the plasma levels of adrenocortical hormones for all subjects, which may correlate with some potential functional SNPs located in the steroid hormone metabolism genes.
Conclusion
In summary, this study conducted a case–control association analysis to investigate the relationship between seven steroid hormone metabolism genes and HAPE in a Chinese Han population. It was found that four SNPs located in HSD3B1, CYP17A1, and CYP11B1, and six haplotypes in CYP17A1 were significantly associated with HAPE. Nevertheless, the relationship between these susceptibility genes and HAPE, as well as the underlying molecular mechanisms, require confirmatory research.
Acknowledgments
We thank those physicians for sample collection, Dr Jiangxi from the Yushu People’s Hospital and Dr Nima from the Yushu Bayi Hospital of Qinghai Province.
Funding
This work was supported by Qinghai Thousand Talents Program, The 2nd Round of “Qinghai 135 High-level Talents Training Project”, Basic Applied Study Foundation of Qinghai (2019-ZJ-7006).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Eichstaedt CA, Mairbäurl H, Song J, et al. Genetic predisposition to high-altitude pulmonary edema. High Alt Med Biol. 2020;21(1):28–36.
2. Li Y, Zhang Y, Zhang Y. Research advances in pathogenesis and prophylactic measures of acute high altitude illness. Respir Med. 2018;145:145–152.
3. Eichstaedt CA, Benjamin N, Grünig E. Genetics of pulmonary hypertension and high-altitude pulmonary edema. J Appl Physiol. 2020;128(5):1432–1438.
4. Yang Y, Xu J, Tang F, et al. NR3C2 gene is associated with susceptibility to high-altitude pulmonary edema in Han Chinese. Wilderness Environ Med. 2018;29(4):488–492.
5. Yang Y, Du H, Li Y, et al. NR3C1 gene polymorphisms are associated with high-altitude pulmonary edema in Han Chinese. J Physiol Anthropol. 2019;38(1):4.
6. Wilcox I, Chan KH, Lattimore JD. Hypoxia and inflammation. N Engl J Med. 2011;364(20):
7. Hartmann G, Tschöp M, Fischer R, et al. High altitude increases circulating interleukin-6, interleukin-1 receptor antagonist and C-reactive protein. Cytokine. 2000;12(3):246–252.
8. Rosenberger P, Schwab JM, Mirakaj V, et al. Hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat Immunol. 2009;10(2):195–202.
9. Estoppey J, Léger B, Vuistiner P, Sartori C, Kayser B. Low- and high-altitude cortisol awakening responses differ between AMS-prone and AMS-resistant mountaineers. High Alt Med Biol. 2019;20(4):344–351.
10. Kumar R, Pasha Q, Khan AP, Gupta V. Renin angiotensin aldosterone system and ACE I/D gene polymorphism in high-altitude pulmonary edema. Aviat Space Environ Med. 2004;75(11):981–983.
11. Schiffer L, Barnard L, Baranowski ES, et al. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: a comprehensive review. J Steroid Biochem Mol Biol. 2019;194:105439.
12. Charu R, Stobdan T, Ram RB, et al. Susceptibility to high altitude pulmonary oedema: role of ACE and ET-1 polymorphisms. Thorax. 2006;61(11):1011–1012.
13. Zheng Y, Huang J. Angiotensin-converting enzyme gene insertion/deletion polymorphism and high-altitude pulmonary edema: an updated meta-analysis. J Renin-Angiotensin-Aldosterone Syst. 2020;21(1):1470320319900039.
14. Zhu X, Bouzekri N, Southam L, et al. Linkage and association analysis of angiotensin I-converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am J Hum Genet. 2001;68(5):1139–1148.
15. Luo Y, Chen Y, Zhang Y, Zhou Q, Gao Y. Association of endothelial nitric oxide synthase (eNOS) G894T polymorphism with high altitude pulmonary edema susceptibility: a meta-analysis. Wilderness Environ Med. 2012;23(3):270–274.
16. Srivastava S, Bhagi S, Kumari B, Chandra K, Sarkar S, Ashraf MZ. Association of polymorphisms in angiotensin and aldosterone synthase genes of the renin-angiotensin-aldosterone system with high-altitude pulmonary edema. J Renin-Angiotensin-Aldosterone Syst. 2012;13(1):155–160.
17. Hotta J, Hanaoka M, Droma Y, Katsuyama Y, Ota M, Kobayashi T. Polymorphisms of renin-angiotensin system genes with high-altitude pulmonary edema in Japanese subjects. Chest. 2004;126(3):825–830.
18. Qi Y, Niu WQ, Zhu TC, et al. Genetic interaction of Hsp70 family genes polymorphisms with high-altitude pulmonary edema among Chinese railway constructors at altitudes exceeding 4000 meters. Clin Chim Acta. 2009;405(1–2):17–22.
19. Hanaoka M, Kubo K, Yamazaki Y, et al. Association of high-altitude pulmonary edema with the major histocompatibility complex. Circulation. 1998;97(12):1124–1128.
20. Gabry AL, Ledoux X, Mozziconacci M, Martin C. High-altitude pulmonary edema at moderate altitude (< 2400 m; 7870 feet): a series of 52 patients. Chest. 2003;123(1):49–53.
21. Jefcoate CR, Lee J. Cholesterol signaling in single cells: lessons from STAR and sm-FISH. J Mol Endocrinol. 2018;60(4):R213–r235.
22. Turcu AF, Auchus RJ. Adrenal steroidogenesis and congenital adrenal hyperplasia. Endocrinol Metab Clin North Am. 2015;44(2):275–296.
23. Swart P, Lombard N, Swart AC, et al. Ovine steroid 17α-hydroxylase cytochrome P450: characteristics of the hydroxylase and lyase activities of the adrenal cortex enzyme. Arch Biochem Biophys. 2003;409(1):145–152.
24. Hattangady NG, Olala LO, Bollag WB, Rainey WE. Acute and chronic regulation of aldosterone production. Mol Cell Endocrinol. 2012;350(2):151–162.
25. Yong Y, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98.
26. Luo Y, Gao W, Chen Y, Liu F, Gao Y. Rare mitochondrial DNA polymorphisms are associated with high altitude pulmonary edema (HAPE) susceptibility in Han Chinese. Wilderness Environ Med. 2012;23(2):128–132.
27. Saxena S, Kumar R, Madan T, Gupta V, Muralidhar K, Sarma PU. Association of polymorphisms in pulmonary surfactant protein A1 and A2 genes with high-altitude pulmonary edema. Chest. 2005;128(3):1611–1619.
28. Stobdan T, Kumar R, Mohammad G, et al. Probable role of beta2-adrenergic receptor gene haplotype in high-altitude pulmonary oedema. Respirology (Carlton, Vic). 2010;15(4):651–658.
29. Zhang Y, Liu T, Wang J, et al. Cellinker: a platform of ligand–receptor interactions for intercellular communication analysis. Bioinformatics. 2021;37(14):2025–2032.
30. Thomas JL, Boswell EL, Scaccia LA, Pletnev V, Umland TC. Identification of key amino acids responsible for the substantially higher affinities of human type 1 3beta-hydroxysteroid dehydrogenase/isomerase (3beta-HSD1) for substrates, coenzymes, and inhibitors relative to human 3beta-HSD2. J Biol Chem. 2005;280(22):21321–21328.
31. Rosmond R, Chagnon M, Bouchard C, Björntorp P. Polymorphism in exon 4 of the human 3 beta-hydroxysteroid dehydrogenase type I gene (HSD3B1) and blood pressure. Biochem Biophys Res Commun. 2002;293(1):629–632.
32. Shimodaira M, Nakayama T, Sato N, et al. Association of HSD3B1 and HSD3B2 gene polymorphisms with essential hypertension, aldosterone level, and left ventricular structure. Eur J Endocrinol. 2010;163(4):671–680.
33. Van Den Akker EL, Koper JW, Boehmer AL, et al. Differential inhibition of 17alpha-hydroxylase and 17,20-lyase activities by three novel missense CYP17 mutations identified in patients with P450c17 deficiency. J Clin Endocrinol Metab. 2002;87(12):5714–5721.
34. Feigelson HS, Shames LS, Pike MC, Coetzee GA, Stanczyk FZ, Henderson BE. Cytochrome P450c17alpha gene (CYP17) polymorphism is associated with serum estrogen and progesterone concentrations. Cancer Res. 1998;58(4):585–587.
35. Li L, Gu ZP, Bo QM, Wang D, Yang XS, Cai GH. Association of CYP17A1 gene −34T/C polymorphism with polycystic ovary syndrome in Han Chinese population. Gynecol Endocrinol. 2015;31(1):40–43.
36. Surekha D, Sailaja K, Rao DN, Padma T, Raghunadharao D, Vishnupriya S. Association of a CYP17 gene polymorphism with development of breast cancer in India. Asian Pac J Cancer Prev. 2010;11(6):1653–1657.
37. Xu J, Yang YZ, Tang F, et al. CYP17A1 and CYP2E1 variants associated with high altitude polycythemia in Tibetans at the Qinghai-Tibetan Plateau. Gene. 2015;566(2):257–263.
38. Caroccia B, Vanderriele PE, Seccia TM, et al. Aldosterone and cortisol synthesis regulation by angiotensin-(1-7) and angiotensin-converting enzyme 2 in the human adrenal cortex. J Hypertens. 2021;39(8):1577–1585.
39. Davies E, Kenyon CJ. CYP11B2 polymorphisms and cardiovascular risk factors. J Hypertens. 2003;21(7):1249–1253.
40. Zhang GX, Wang BJ, Ouyang JZ, et al. Polymorphisms in CYP11B2 and CYP11B1 genes associated with primary hyperaldosteronism. Hypertension Res. 2010;33(5):478–484.
41. Chu M, Wu S, Wang W, et al. miRNA sequencing reveals miRNA-4508 from peripheral blood lymphocytes as potential diagnostic biomarker for silica-related pulmonary fibrosis: a multistage study. Respirology (Carlton, Vic). 2020;25(5):511–517.
42. Hobbs BD, de Jong K, Lamontagne M, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49(3):426–432.
43. Hobbs BD, Putman RK, Araki T, et al. Overlap of genetic risk between interstitial lung abnormalities and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2019;200(11):1402–1413.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
