Back to Journals » OncoTargets and Therapy » Volume 9
Association between the Cyclin D1 G870A polymorphism and the susceptibility to and prognosis of upper aerodigestive tract squamous cell carcinomas: an updated meta-analysis
Authors Meng Y, Zhang C, Zhou X
Received 17 August 2015
Accepted for publication 14 December 2015
Published 19 January 2016 Volume 2016:9 Pages 367—376
DOI https://doi.org/10.2147/OTT.S94635
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Yichen Meng,* Chenglin Zhang,* Xuhui Zhou
Changzheng Hospital, Second Affiliated Hospital of Second Military Medical University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Purpose: Several publications have investigated the association between the Cyclin D1 G to A substitution at nucleotide 870 (CCND1 G870A) polymorphism and squamous cell carcinoma (SCC) of the upper aerodigestive tract (UADT), but their conclusions still remain controversial. We conducted a meta-analysis to precisely evaluate this association.
Patients and methods: We electronically searched the Chinese National Knowledge Infrastructure, PubMed, and Embase (up to January 2015) databases for case–control studies on the association between the CCND1 G870A polymorphism and SCC of the UADT, and 23 studies were included in total.
Results: The meta-analysis results showed that there was a significant association between the CCND1 G870A polymorphism and the risk of SCC of the UADT (AA vs GG: odds ratio [OR] =1.33, 95% confidence interval [CI] =1.01–1.74, P<0.001 for heterogeneity; GA/AA vs GG: OR =1.24, 95% CI =1.01–1.51, P<0.001 for heterogeneity; AA vs GA/GG: OR =1.16, 95% CI =0.97–1.39, P<0.001 for heterogeneity; allele A vs allele G: OR =1.14, 95% CI =1.00–1.30, P<0.001 for heterogeneity; GA vs GG: OR =1.18, 95% CI =0.98–1.42, P<0.001 for heterogeneity). However, when analyzing prognosis, allele G was a potential risk factor for poor tumor differentiation (AA vs GA/GG: OR =2.60, 95% CI =1.15–5.86, P=0.836 for heterogeneity) and reduced disease-free intervals (OR =2.08, 95% CI =1.17–3.69, P=0.134 for heterogeneity). In the subgroup analysis, the cancer susceptibility of Asian groups, population-based control groups, nasopharyngeal cancer groups, and esophageal SCC groups were more likely to be affected by the CCND1 G870A polymorphism. No significant publication bias was found in our analysis (P=0.961 for Egger’s test and P=0.245 for Begg’s test).
Conclusion: The results of the present meta-analysis suggest that the variant CCND1 870A allele might confer an elevated risk of SCC of the UADT, particularly among Asians and individuals who have esophageal or nasopharyngeal cancers. Moreover, the CCND1 870A allele might also confer better tumor differentiation grades and longer disease-free intervals.
Keywords: CCND1 G870A polymorphism, squamous cell carcinoma, predictive value, screen
Introduction
Cancers of the upper aerodigestive tract (UADT), including the oral cavity, pharynx, larynx, and esophagus, are the fourth most common type of cancer and account for approximately 5%–8% of all malignancies worldwide.1,2 Among UADT cancers, squamous cell carcinomas (SCCs) constitute the most abundant histologic type.3 Although the detailed mechanisms of the carcinogenesis of SCC of the UADT still remain ambiguous, several studies suggest that genetic factors play an important role in this process. Thus, it is of great importance to identify potential genetic factors to help screen the susceptible population.
Cyclin D1 (CCND1) is a key regulator in the transition from the G1 phase to the S phase during cell cycle progression.4 Several publications have reported that CCND1 gene amplification is involved in the pathogenesis of a variety of cancers, including breast, prostate, esophagus, and lung cancer.5–8 The CCND1 gene has an important polymorphism (rs603965) at codon 242 in exon 4 that results in a silent G to A substitution at nucleotide 870 (G870A).9 The shift from allele G (wild-type gene) to allele A (variant gene) leads to the expression of an altered protein that has a longer half-life and thus enables cells to more easily bypass the G1/S cell cycle checkpoint.10,11 Additionally, evidence has shown that the carriers of the AA genotype are more likely to have numerous cancers, including brain, liver, and breast cancer.12–14
The association between the CCND1 G870A polymorphism and SCC of the UADT has been investigated by different authors, but their conclusions still remain controversial. A recent meta-analysis by Wen et al15 showed a possible relationship between the CCND1 G870A polymorphism and esophageal cancer risk, which was contrary to the results found from an earlier meta-analysis conducted by Cai et al.16 We consider that this difference results from sample sizes. Another meta-analysis by Tang et al17 pooled head and neck cancers of different histological types together, preventing a detailed subgroup analysis; thus, they obtained unconvincing results with high heterogeneity. These studies did not assess the influence of the CCND1 G870A polymorphism on the prognosis of UADT cancers. In addition, four additional studies reporting the association of the CCND1 G870A polymorphism with UADT SSC have been published. Therefore, we performed this meta-analysis including all relevant published studies to provide complementary and updated information on the relationship between the CCND1 G870A polymorphism and the susceptibility and prognosis of UADT SCC.
Patients and methods
Literature search strategy
We performed an electronic search of the Chinese National Knowledge Infrastructure, PubMed, and Embase (up to January 2015) databases for case–control studies on the association between the CCND1 G870A polymorphism and SCC of the UADT. The following terms were used in combination: Cyclin D1, CCND1, polymorphism, squamous cell carcinoma, oral cancer, laryngeal cancer, pharyngeal cancer, esophageal cancer, and case–control study. Manual searches were also conducted to identify relevant publications from the citation lists of the included articles.
Inclusion and exclusion criteria
Studies were included if they met the following criteria: 1) a case–control study design concerning the relationship between the CCND1 G870A polymorphism and SCC of the UADT; 2) SCC confirmed via histopathological evidence; 3) tumor sites located in the UADT (including oral cavity, larynx, pharynx, and esophagus); 4) reported odds ratios (ORs) with a 95% confidence interval (CI) (or crude data that allowed the 95% CI to be calculated); and 5) published in English or Chinese.
Studies were excluded if they met the following criteria: 1) case series, case reports, reviews, and descriptive studies; 2) reports with a small number of participants (fewer than 20 patients); 3) crude data were not given or ORs could not be calculated; and 4) not published in English or Chinese.
Data extraction
Two authors independently read the full texts and extracted relevant data from each publication. The following data were extracted: the first author, year of publication, country, ethnicity of subjects, type of controls, genotyping methods, Hardy–Weinberg equilibrium (HWE) in controls, and genotype distributions of cases and controls. After the first extraction, the data were rechecked by two authors, and disagreements during the extraction were resolved by consensus.
Statistical analysis
Statistical analyses were performed using Stata software version 12.0 (Stata Corporation, College Station, TX, USA). We pooled the ORs with 95% CIs to analyze the association between the CCND1 G870A polymorphism and susceptibility to SCC of the UADT. Four genetic models were evaluated, including the allele-contrast model (A vs G), the codominant model (AA vs GG and GA vs GG), the dominant model (GA/AA vs GG), and the recessive model (AA versus GA/GG) (GG wild-type homozygotes, GA heterozygotes, and AA variant homozygotes). The I-squared (I2) statistic was used to evaluate statistical heterogeneity among studies, and I2 values of <25% and >50% reflected low and high heterogeneity, respectively.18,19 In our analysis, both the fixed-effects model and random-effects model were used. The random-effects model was used only when the P-values of the heterogeneity test were less than 0.1. In addition, funnel plots and Egger’s linear regression test were used to detect publication bias.20
Results
A total of 231 articles were identified after searching the Chinese National Knowledge Infrastructure, PubMed, and Embase databases. After the exclusion of 128 articles that were irrelevant to our research, 103 remained. In total, 32 articles were retrieved for full-text review, and finally, nine of them were excluded because the crude data could not be extracted. Therefore, 23 studies matched the inclusion criteria and were chosen for the following meta-analysis (Figure 1).21–43
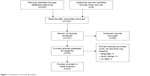  | Figure 1 Flowchart of screening studies. |
These studies are dated from 1998 to 2014 with case–control designs and included a total of 11,542 subjects, of whom 5,338 (46.2%) were histopathologically diagnosed with SCC of the UADT. Detailed information extracted from the included publications is provided in Table 1. Of the 23 included studies, there were ten groups of Caucasians,21–23,25,28,31,32,34,38,43 ten groups of Asians,21,26,27,29,35–37,39–42 and three groups of mixed ethnicities.24,30,33 Among the 23 included studies, the classic polymerase chain reaction-restriction fragment length polymorphism assay was the most frequently used method to genotype the CCND1 G870A polymorphism. Genotyping distributions of the control populations were consistent with HWE in all studies except for those in two studies. Lin et al29 did not report the precise distribution of genotypes GG and GA in their study and combined them as GG + GA. Seven studies were hospital-based24,25,31,33,35,37,40 and 16 were population-based.21–23,26–30,32,34,36,38,39,41–43
Overall, the meta-analysis results suggested that there was a significant association between the CCND1 G870A polymorphism and risk of SCC of the UADT under the codominant model (AA vs GG: OR =1.33, 95% CI =1.01–1.74, P<0.001 for heterogeneity), the dominant model (GA/AA vs GG: OR =1.24, 95% CI =1.01–1.51, P<0.001 for heterogeneity), and the recessive model (AA vs GA/GG: OR =1.16, 95% CI =0.97–1.39, P<0.001 for heterogeneity). Borderline elevated risks were also found in the allele-contrast model (allele A vs allele G: OR =1.14, 95% CI =1.00–1.30, P<0.001 for heterogeneity) and the codominant model (GA vs GG: OR =1.18, 95% CI =0.98–1.42, P<0.001 for heterogeneity).
We also examined associations between the CCND1 G870A polymorphism and the patient prognosis of SCC, including tumor differentiation, tumor size, lymph node involvement, recurrence, and disease-free interval. Differentiation was graded as moderate (G0–2) or poor (G3). Tumor size was graded as large (T3 + T4) or small (T1 + T2). Lymph node involvement and recurrence were graded as positive or negative. Tumor differentiation was reported in three studies,25,31,34 and the meta-analysis results showed that patients with AA genotypes were less likely to have poorly differentiated tumors (AA vs GA/GG: OR =2.60, 95% CI =1.15–5.86, P=0.836 for heterogeneity, Figure 2). However, we did not find any significant association with AA genotypes for lymph node involvement, tumor size, and recurrence (AA vs GA/GG: OR =0.85, 95% CI =0.46–1.57, P=0.03 for heterogeneity, Figure 3; OR =0.97, 95% CI =0.71–1.32, P=0.801 for heterogeneity, Figure 4; OR =0.60, 95% CI =0.36–1.01, P=0.328 for heterogeneity, Figure 5, respectively). The survival or disease-free interval was extracted as the hazard ratio with a 95% CI. Two studies reported the disease-free interval,29,31 and two studies reported the survival.30,40 Pooled data demonstrated that allele G was a potential risk factor for reductions in the disease-free interval (OR =2.08, 95% CI =1.17–3.69, P=0.134 for heterogeneity, Figure 6), but this effect was not found for the survival interval (OR =0.91, 95% CI =0.37–2.23, P=0.074 for heterogeneity, Figure 6).
Subgroup and sensitivity analysis
Considering the strong evidence of heterogeneity, subgroup analyses stratified by ethnicity (Caucasian, Asian, or Mixed), type of control (hospital-based or population-based), smoking status (ever smoking or never smoking), and tumor site (oral cavity, larynx, pharynx, nasopharynx, or esophagus) were conducted. The main results of the subgroup analysis are shown in Table 2. In the subgroup stratified by ethnicity, a significantly increased cancer risk was found among Asians under all genetic models. In the subgroup by source of controls, population-based studies suggested an elevated risk of SCC of the UADT. The subgroup analysis of the tumor site revealed that the risks of nasopharyngeal cancer and esophageal SCC were more likely to be affected by the CCND1 G870A polymorphism. No significant risk effect was found in the subgroup analysis by smoking status.
  | Table 2 Subgroup analysis of the included studies |
Sensitivity analysis was performed by excluding the two studies that deviated from HWE.24,37 Additionally, the meta-analysis results in any genetic model were not statistically altered (data not shown). In addition, after excluding each single study one by one, the pooled results were not significantly attenuated, which confirmed the robustness of the data (data not shown).
Publication bias
A funnel plot of log OR vs standard error was constructed for the recessive model, under which all 23 studies were covered (Figure 7). Visual inspection of the funnel plots did not show any remarkable asymmetry. In addition, Egger’s and Begg’s tests showed no significant publication bias (P=0.961 for Egger’s test and P=0.245 for Begg’s test, respectively).
Discussion
We were the first to conduct a meta-analysis involving 5,338 cases and 6,204 controls to evaluate the relationship between the CCND1 G870A polymorphism and susceptibility to SCC of the UADT. Furthermore, the influence of the polymorphism on the prognosis of SCC was also investigated. Finally, based on a total of 23 studies, our meta-analysis suggested that allele A of the CCND1 G870A polymorphism is associated with SCC development of the UADT, especially among Asian groups, groups with population-based controls, and groups of nasopharyngeal carcinoma and esophageal carcinoma. However, homozygous AA appeared to be a protective factor, as patients with AA genotypes had increased disease-free intervals and a higher degree of tumor differentiation.
The CCND1 gene, which is located on chromosome 11q13, is polymorphic with a common G870A in the conserved splice donor region of exon 4 of the gene. The CCND1 G870A polymorphism has been reported to have a promising role in predicting carcinogenesis of SCC, but this conclusion is still debated. Matthias et al31 first investigated the influence of the CCND1 G870A polymorphism on the susceptibility to SCC of the head and neck (SCCHN), and no significant correlation was found. However, they demonstrated that the GG genotype was associated with poor differentiation and a shortened time for tumor recurrence. Similarly, Monteiro et al32 did not find any evident relationship between the CCND1 G870A polymorphism and laryngeal SCC susceptibility. Their conclusion differed from that of Matthias et al31 in that their data did not support an association between the CCND1 genotype and tumor differentiation grade or recurrent disease. Marsit et al30 found that the homozygous AA genotype was associated with a 1.5-fold elevated risk of SCCHN, which was in accordance with findings reported by Zheng et al,43 but was opposite to that reported by Holley et al,25 who found a significantly elevated risk (OR =3.37, 95% CI =1.61–9.80) for oral SCCs in subjects possessing the GG genotype. We considered that these discrepancies were due to several reasons: first, these studies all had relatively small sample sizes, which provided limited statistical power; second, these studies were conducted in different countries and included samples that may have had different genetic backgrounds; and third, gene polymorphisms may have different biological significances in various cancers, but some studies grouped tumors from various anatomic sites together. Thus, meta-analysis was required. A recent meta-analysis indicated that there was an overall lack of association between the CCND1 G870A polymorphism and head and neck cancer risk under all five genetic models, and no significant association was found in the subgroup analysis.44 Cases in that study were diagnosed with different pathological types of cancers, which were located in the head and neck. In the current study, publications targeting SCC specifically originating from the UADT were collected, and subgroups were stratified by ethnicity, source of controls, smoking status, and tumor site. In the subgroup analysis of ethnicity, a remarkable association between CCND1 allele A and elevated cancer risk was observed among Asians but not other ethnicities. This race-prevalent phenomenon, which might be due to the specific ethnic chromosomal aberrations and different genetic backgrounds, was also observed in lung cancer.45 In the subgroup analysis of the source of controls, allele A was found to be associated with increased cancer risk in the population-based subgroup. Population-based controls were more likely to represent the general population compared with hospital-based controls. As for hospital-based controls, those with other diseases may be of higher risk in terms of exposure status, such as alcohol consumption, which may potentially influence the disease studied. Thus, selection bias from hospital-based studies might attenuate the overall results of the meta-analysis. As a result, investigators should use proper controls to reduce possible selection bias in future studies. Monteiro et al32 reported that tobacco exposure contributed to CCND1 dysregulation. In our meta-analysis, a total of seven studies reported the interaction between smoking and the CCND1 G870A polymorphism in SCC susceptibility,25,26,36,37,39,42,43 but only two of them provided detailed genotype distributions of subgroups concerning smoking status.37,43 Pooled data of these two studies failed to find any significant correlation between the CCND1 polymorphism and smoking. Another three studies on esophageal SCC revealed that smoking patients had a significantly higher allele A frequency compared to healthy smokers, but this disparity was not found among nonsmokers.26,39,42 Consequently, our subgroup analysis results should be interpreted with caution because of the limited number and small sample sizes of the included studies. Wen et al’s15 meta-analysis suggested that the CCND1 G870A polymorphism presents the potential for elevated risk in the development of esophageal malignancies, including both adenocarcinomas and SCCs. In the current study, the subgroup analysis of the tumor site showed that allele A was related to an elevated risk of nasopharyngeal and esophageal SCCs. However, only one study regarding nasopharyngeal carcinoma was included and the statistical power was limited.
Further analysis of the association between the CCND1 G870A polymorphism and the prognosis of SCC of the UADT was carried out. Patients with the AA genotype were more likely to have well-differentiated tumors, and allele G was a potential risk factor for reduced disease-free interval. Holley et al46 reported that the GG genotype was associated with poorly differentiated tumors, which was inconsistent with our findings. However, when both CCND1 genotypes were included in a model with age and sex, CCND1 GG was no longer significantly associated (P=0.76, OR =1.17, 95% CI =0.43–3.23). Thus, more data were required to validate the association between the CCND1 G870A polymorphism and the prognosis of SCC of the UADT.
Our study still had some limitations. First, although Egger’s test did not indicate any statistically significant differences in our study, publication bias still cannot be ignored because only publications in English and Chinese were included in our analysis. There is a possibility that we missed certain eligible studies. Second, the data pooled in our analysis were unadjusted; however, carcinogenesis is an intricate process influenced by many factors other than genes. Effects of the gene–gene and gene–environment interactions in the development of SCC of the UADT could not be evaluated. Third, the sample sizes of included studies were relatively small, which limited the power of our results.
Conclusion
Despite these limitations, the results of the present meta-analysis suggest that the variant CCND1 870A allele might confer an elevated risk of SCC of the UADT, particularly among Asians and individuals who have esophageal or nasopharyngeal cancers. Moreover, the CCND1 870A allele might also promote higher tumor differentiation grades and longer disease-free intervals. These findings may help to understand the role of the CCND1 G870A polymorphism in the etiology of SCC of the UADT. In the future, large case–control studies that consider gene–gene and gene–environment interactions are warranted to verify these findings.
Disclosure
The authors report no conflicts of interest in this work.
References
Delahaye-Sourdeix M, Anantharaman D, Timofeeva MN, et al. A rare truncating BRCA2 variant and genetic susceptibility to upper aerodigestive tract cancer. J Natl Cancer Inst. 2015;107(5):djv037. | ||
Steffen A, Bergmann MM, Sanchez MJ, et al. Meat and heme iron intake and risk of squamous cell carcinoma of the upper aero-digestive tract in the European Prospective Investigation into Cancer and Nutrition (EPIC). Cancer Epidemiol Biomarkers Prev. 2012;21(12):2138–2148. | ||
Cooper JS, Porter K, Mallin K, et al. National Cancer Database report on cancer of the head and neck: 10-year update. Head Neck. 2009;31(6):748–758. | ||
Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. | ||
Jovanovic IP, Radosavljevic GD, Simovic-Markovic BJ, et al. Clinical significance of Cyclin D1, FGF3 and p21 protein expression in laryngeal squamous cell carcinoma. J BUON. 2014;19(4):944–952. | ||
Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–572. | ||
Noel EE, Yeste-Velasco M, Mao X, et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am J Pathol. 2010;176(6):2607–2615. | ||
Ravikumar G, Ananthamurthy A. Cyclin D1 expression in ductal carcinoma of the breast and its correlation with other prognostic parameters. J Cancer Res Ther. 2014;10(3):671–675. | ||
Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. | ||
Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. | ||
Wei M, Zhu L, Li Y, et al. Knocking down cyclin D1b inhibits breast cancer cell growth and suppresses tumor development in a breast cancer model. Cancer Sci. 2011;102(8):1537–1544. | ||
Akkiz H, Bayram S, Bekar A, Akgollu E, Ozdil B. Cyclin D1 G870A polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: case–control study. Cancer Epidemiol. 2010;34(3):298–302. | ||
Canbay E, Eraltan IY, Cercel A, et al. CCND1 and CDKN1B polymorphisms and risk of breast cancer. Anticancer Res. 2010;30(7):3093–3098. | ||
Zeybek U, Yaylim I, Ozkan NE, et al. Cyclin D1 gene G870A variants and primary brain tumors. Asian Pac J Cancer Prev. 2013;14(7):4101–4106. | ||
Wen L, Hu YY, Yang GL, Liu DX. CCND1 G870A polymorphism contributes to the risk of esophageal cancer: an updated systematic review and cumulative meta-analysis. Biomed Rep. 2014;2(4):549–554. | ||
Cai W, Wang ZT, Zhong J, Zhang Y. Lack of association between Cyclin D1 gene G870A polymorphism and esophageal cancer: evidence from a meta-analysis. Genet Mol Res. 2013;12(4):6636–6645. | ||
Tang C, Wang Z, Yu J, Wu Y, Zhu Z, Chen N. CCND1 G870A polymorphism and risk for head and neck cancer: a meta-analysis. Med Oncol. 2011;28(4):1319–1324. | ||
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Akbari MR, Malekzadeh R, Shakeri R, et al. Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 2009;69(20):7994–8000. | ||
Atac A, Riecke B, Assaf AT, et al. Lack of evidence for predictive and prognostic value of cyclin D1 gene polymorphism CCND1 G870A for oral squamous cell carcinoma. Anticancer Res. 2014;34(1):235–238. | ||
Djansugurova LB, Perfilyeva AV, Zhunusova GS, Djantaeva KB, Iksan OA, Khussainova EM. The determination of genetic markers of age-related cancer pathologies in populations from Kazakhstan. Front Genet. 2013;4:70. | ||
Gomes CC, Drummond SN, Guimaraes AL, Andrade CI, Mesquita RA, Gomez RS. P21/WAF1 and cyclin D1 variants and oral squamous cell carcinoma. J Oral Pathol Med. 2008;37(3):151–156. | ||
Holley SL, Matthias C, Jahnke V, Fryer AA, Strange RC, Hoban PR. Association of cyclin D1 polymorphism with increased susceptibility to oral squamous cell carcinoma. Oral Oncol. 2005;41(2):156–160. | ||
Hussain S, Yuvaraj M, Thakur N, et al. Association of cyclin D1 gene polymorphisms with risk of esophageal squamous cell carcinoma in Kashmir valley: a high risk area. Mol Carcinog. 2011;50(7):487–498. | ||
Jang Y, Lu SA, Chen ZP, et al. Genetic polymorphisms of CCND1 and PTEN in progression of esophageal squamous carcinoma. Genet Mol Res. 2013;12(4):6685–6691. | ||
Kurmano BK, Djansugurova L, Bersimba R, et al. TP53 Arg72Pro and CCND1 A870G polymorphisms and esophageal cancer risk. J Life Sci. 2010;4(6):16–20. | ||
Lin YC, Yeh CJ, Wang LH, Lee CW, Chen CH. The effect of CCND1 +870A>G and VEGF +936C>T polymorphisms on oral cancer development and disease-free survival in a Taiwan population. Oral Oncol. 2012;48(6):535–540. | ||
Marsit CJ, Black CC, Posner MR, Kelsey KT. A genotype–phenotype examination of cyclin D1 on risk and outcome of squamous cell carcinoma of the head and neck. Clin Cancer Res. 2008;14(8):2371–2377. | ||
Matthias C, Branigan K, Jahnke V, et al. Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998;4(10):2411–2418. | ||
Monteiro E, Varzim G, Pires AM, Teixeira M, Lopes C. Cyclin D1 A870G polymorphism and amplification in laryngeal squamous cell carcinoma: implications of tumor localization and tobacco exposure. Cancer Detect Prev. 2004;28(4):237–243. | ||
Nishimoto IN, Pinheiro NA, Rogatto SR, et al. Cyclin D1 gene polymorphism as a risk factor for squamous cell carcinoma of the upper aerodigestive system in non-alcoholics. Oral Oncol. 2004;40(6):604–610. | ||
Rydzanicz M, Golusinski P, Mielcarek-Kuchta D, Golusinski W, Szyfter K. Cyclin D1 gene (CCND1) polymorphism and the risk of squamous cell carcinoma of the larynx. Eur Arch Otorhinolaryngol. 2006;263(1):43–48. | ||
Sathyan KM, Nalinakumari KR, Abraham T, Kannan S. Influence of single nucleotide polymorphisms in H-Ras and cyclin D1 genes on oral cancer susceptibility. Oral Oncol. 2006;42(6):607–613. | ||
Sui J, Gao W, Li XJ, et al. Cyclin D1 gene G870A polymorphism and susceptibility to nasopharyngeal carcinoma. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2009;44(3):227–231. | ||
Tsai MH, Tsai CW, Tsou YA, Hua CH, Hsu CF, Bau DT. Significant association of cyclin D1 single nucleotide polymorphisms with oral cancer in Taiwan. Anticancer Res. 2011;31(1):227–231. | ||
Verim A, Ozkan N, Turan S, et al. Association of the Cylin D1 G870A polymorphism with laryngeal cancer: are they really related? Asian Pac J Cancer Prev. 2013;14(12):7629–7634. | ||
Wang R, Zhang JH, Li Y, Wen DG, He M, Wei LZ. The association of cyclin D1 (A870G) polymorphism with susceptibility to esophageal and cardiac cancer in north Chinese population. Zhonghua Yi Xue Za Zhi. 2003;83(12):1089–1092. | ||
Wong YK, Lin SC, Chang CS, et al. Cyclin D1 genotype in areca-associated oral squamous cell carcinoma. J Oral Pathol Med. 2003;32(5):265–270. | ||
Yu C, Lu W, Tan W, et al. Lack of association between CCND1 G870A polymorphism and risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2003;12(2):176. | ||
Zhang J, Li Y, Wang R, et al. Association of cyclin D1 (G870A) polymorphism with susceptibility to esophageal and gastric cardiac carcinoma in a northern Chinese population. Int J Cancer. 2003;105(2):281–284. | ||
Zheng Y, Shen H, Sturgis EM, et al. Cyclin D1 polymorphism and risk for squamous cell carcinoma of the head and neck: a case–control study. Carcinogenesis. 2001;22(8):1195–1199. | ||
Lin H, Fang L, Lin D. Association of cyclin D1 variants with head and neck cancer susceptibility: evidence from a meta-analysis. Asian Pac J Cancer Prev. 2014;15(14):5645–5651. | ||
Broet P, Dalmasso C, Tan EH, et al. Genomic profiles specific to patient ethnicity in lung adenocarcinoma. Clin Cancer Res. 2011;17(11):3542–3550. | ||
Holley SL, Parkes G, Matthias C, et al. Cyclin D1 polymorphism and expression in patients with squamous cell carcinoma of the head and neck. Am J Pathol. 2001;159(5):1917–1924. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







