Back to Journals » OncoTargets and Therapy » Volume 8
Association between GSTP1 polymorphisms and prognosis of osteosarcoma in patients treated with chemotherapy: a meta-analysis
Authors Pu F , Chen F, Chen S, Wang B, Liu J, Shao Z
Received 17 January 2015
Accepted for publication 3 June 2015
Published 23 July 2015 Volume 2015:8 Pages 1835—1842
DOI https://doi.org/10.2147/OTT.S81115
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Feifei Pu,1,* Fengxia Chen,2,* Songfeng Chen,1 Baichuan Wang,1 Jianxiang Liu,1 Zengwu Shao1
1Department of Orthopedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China; 2Department of Medical Oncology, General Hospital of The Yangtze River Shipping, Wuhan, Hubei, People’s Republic of China
*These authors contributed equally to this work
Objective: The aim of this study was to evaluate the relationship between GSTP1 polymorphisms and prognosis of osteosarcoma in patients treated with chemotherapy, by performing a meta-analysis.
Methods: The studies of effects of GSTP1 gene polymorphisms on osteosarcoma survival after chemotherapy were collected. STATA (version 12.0) was used to perform data synthesis.
Results: Six studies involving 898 participants were included. A meta-analysis was performed on studies in GSTP1 313A>G(rs1695) assessing the association between tumor response and the polymorphisms in GSTP1 (AA vs AG, AA vs GG), the pooled odds ratios (ORs) were 2.06 (95% confidence interval [CI]: 1.48–2.86, P=0.628, I2=0.0%). There was significant association between the polymorphisms in GSTP1 (AA vs AG, AA vs GG) and the events that happened, the pooled ORs were 1.86 (95% CI: 1.14–3.06, P=0.034, I2=58.6%), and there was significant association between the polymorphisms in GSTP1 (AA vs AG, AA vs GG) and survival times (overall survival and progression-free survival) in osteosarcoma patients treated with chemotherapy, and the pooled ORs were 2.14 (95% CI: 1.51–3.04, P=0.675, I2=0.0%) and 2.77 (95% CI: 1.56–4.91, P=0.347, I2=9.3%), respectively. Two studies assessed the association of polymorphisms in GSTP1 I105V (IIe/IIe vs IIe/Val, IIe/IIe vs Val/Val) with overall survival in human osteosarcoma. The pooled ORs were 1.20 (95% CI: 0.64–2.27, P=0.010, I2=73.5%). The study showed an insignificant difference in overall survival for the polymorphisms in GSTP1 (IIe/IIe vs IIe/Val, IIe/IIe vs Val/Val).
Conclusion: This meta-analysis indicated that GSTP1 polymorphisms might influence osteosarcoma risk and suggests that GSTP1 polymorphisms may be an important risk factor for osteosarcoma.
Keywords: GSTP1, polymorphisms, osteosarcoma, chemotherapy, prognosis, meta-analysis
Introduction
Osteosarcoma derives from primitive bone-forming mesenchymal cells, and it is the most common malignant tumor of bones and one of the leading cause of death from cancer in children and adolescents.1 The real mechanism of developing osteosarcoma is still not well understood, but its genesis and progression may be regulated by genetic factors.2 Standard treatment of osteosarcoma involves neoadjuvant therapy before definitive resection of the primary tumor, followed by multiple chemotherapeutic agents after operation.3 Nevertheless, more than 40% patients have a poor tumor response to chemotherapy, and the estimated survival is approximately 45%–55%,4 and the 5-year overall relapse-free survival rate is approximately 65%.5 According to biomarkers, individualized chemotherapy may improve the patients’ tumor response to chemotherapy and the clinical outcome. Therefore, fully understanding the role of pharmacogenetics could help establish individualized chemotherapy for patients to benefit more from chemotherapy to prolong their life. In recent years, several common gene polymorphisms, including in GSTP1, have been identified to be associated with osteosarcoma.6
Glutathione S-transferases (GSTs) are a family of cytosolic enzymes, which are involved in metabolizing chemotherapeutic agents and play an important role in the detoxification of various exogenous and endogenous reactive species.7,8 Glutathione S-transferase p1 (GSTP1) belongs to the pi class and encodes a glutathione S-transferase. This gene is a polymorphic gene encoding differently active, functional GSTP1 proteins in xenobiotic metabolism.9 GSTP1 plays a role in susceptibility to some cancers, such as breast cancer and osteosarcoma.6,10 The genetic polymorphisms could affect drug metabolizers and transporters, which could affect the plasma concentration of chemotherapy drugs and influence the effectiveness of chemotherapy.
Numerous studies have reported GSTP1 polymorphisms are associated with prognosis of osteosarcoma patients treated with chemotherapy, especially GSTP1 313A>G and GSTP1 I105V, but the results are inconclusive, which may be due to the limitation of the small sample size in each of published studies. Therefore, we performed a meta-analysis of the published studies to estimate the association more accurately.
Materials and methods
Ethics statement
The study was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology and conforms with the provisions of the Declaration of Helsinki.
Search strategies
We searched for non-English and English articles included in PubMed, SCI, Elsevier, and CNKI database. Search terms included the following key words: glutathione S-transferase p1, GSTP1, polymorphisms, osteosarcoma, and chemotherapy. Studies up to October 2014 were included. There was no restriction on time period, population, language, or type of study. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. The computerized search was supplemented by a manual search of the bibliographies of all retrieved articles by two independent reviewers.
Inclusion/exclusion criteria of literature
The studies were included if they satisfied the following criteria:
- The study design was case-control;
- They had information on the relationship between GSTP1 polymorphisms and osteosarcoma risk or clinical outcome after chemotherapy;
- The papers provided the size of the samples, distribution of alleles, genotypes, or other informations that could help us estimate an odds ratio and 95% confidence interval.
The studies were excluded if one of the following existed:
- If they were review articles and studies that contained overlapping data;
- If they did not offer the source of cases and controls or other essential information;
- If more than one study from the same group occurred, we only used the most recent study or the study where complete data was available.
Statistical analysis
The following information was extracted from each eligible study by two investigators independently with the standard protocol: the name of first author, year of publication, country of origin, ethnicity of the population studied, the sample size of cases and controls, genotyping methods and allele and genotype distribution in cases and controls, tumor response after chemotherapy, overall survival, progression-free survival, event-free survival, and events after chemotherapy. Different ethnicities of the population were categorized as Caucasians, Asian. To ensure the accuracy of the extracted information, two investigators checked the data extraction results and reached consensus on all of the data extracted. If different results were generated, they would check the data again and have a discussion to come to an agreement. The results were reviewed by a third investigator and any disagreement was resolved by discussion.
Risk ratio (RR) and a 95% CI were used for presenting the statistical results for dichotomous outcomes. Weighted mean difference (WMD) and a 95% CI were employed for presenting the statistical results for continuous outcomes. Mantel–Haenszel analysis was utilized for dichotomous variables, and inverse variance method was used for continuous variables.11 The statistical significance was set at P<0.05.
The difference was considered to be statistically significant if a P-value was less than 0.10 and was also quantitatively assessed by using the value of I-square (I2<25%, no heterogeneity; I2 between 25% and 50%, moderate heterogeneity; and I2>50%, high heterogeneity).12 If I2<50% or P>0.10, it shows that the studies were homogeneous or slightly heterogeneous, and we used the fix effects model to combine the effect size. If I2>50% or P<0.10, indicating that the studies were moderately or highly heterogeneous, and we employed the random effects model to combine the effect size.13 Statistical calculations were performed by using STATA version 12.0 (StataCorp LP, College Station, TX, USA).
Results
Search results and features of studies included
The initial search was independently executed by two reviewers, and 25 articles were preliminarily selected based on the research criteria. After screening by title and abstract, in accordance with inclusion/exclusion criteria, 19 studies were excluded. After a thorough discussion between the two reviewers, six articles were found to be related to this study. The 36 articles were then subjected to a second stage of review. Finally, a total of six studies were included for the meta-analysis. The detailed steps of the literature search are shown in Figure 1. Six studies,14–19 covering data from a total of 898 participants, were included in this review. The information about the authors, publication year, national sources, ethnicity, method, genotype, and outcome parameter of each study are listed in Table 1. There were five studies in Asians and only one study in Caucasians. The genotype distribution in the controls of all the studies included in our meta-analysis was consistent with the Hardy–Weinberg equilibrium (HWE). Information on gene and the sample size, OR estimation, and 95% CI of each study are given in Table 2.
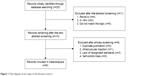  | Figure 1 Flow diagram of the steps in the literature search. |
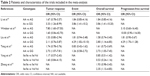  | Table 2 Patients and characteristics of the trials included in the meta-analysis |
Result of meta-analysis
A meta-analysis was performed on studies relating to GSTP1 313A>G(rs1695) to assess the association between tumor response in osteosarcoma patients treated with chemotherapy and the polymorphisms in GSTP1 (AA vs AG, AA vs GG). The pooled ORs were 2.06 (95% CI: 1.48–2.86, P=0.628, I2=0.0%) (Figure 2). There was significant association between the polymorphisms in GSTP1 (AA vs AG, AA vs GG) and the events that happen in osteosarcoma patients treated with chemotherapy, and the pooled ORs were 1.86 (95% CI: 1.14–3.06, P=0.034, I2=58.6%) (Figure 3). There was significant association between the polymorphisms in GSTP1 (AA vs AG, AA vs GG) and survival times (overall survival and progression-free survival) in osteosarcoma patients treated with chemotherapy, the pooled ORs were 2.14 (95% CI: 1.51–3.04, P=0.675, I2=0.0%) and 2.77(95% CI: 1.56–4.91, P=0.347, I2=9.3%), respectively (Figures 4 and 5).
Two studies assessed the association of polymorphisms in GSTP1 I105V (IIe/IIe vs IIe/Val, IIe/IIe vs Val/Val) with overall survival in human osteosarcoma. The pooled ORs were 1.20 (95% CI: 0.64–2.27, P=0.010, I2=73.5%) (Figure 6). The study showed an insignificant difference in overall survival for the polymorphisms in GSTP1 (IIe/IIe vs IIe/Val, IIe/IIe vs Val/Val).
Publication bias
We did not draw funnel plots to demonstrate publication bias, because the number of studies included was comparatively small.
Discussion
The GST gene is a target gene which can regulate human metabolism, the immune and endocrine systems, and has a close relationship with the occurrence of osteosarcoma. It has been confirmed that different GST genes have different conversion efficiencies and enantioselectivities for various toxins which may lead to cancer.8,20 GSTP1 is the most important member of GST family, which takes part in the detoxification of electrophilic compounds by glutathione conjugation.21 GSTP1 polymorphism links to enzymatic activity, and the polymorphism may confer susceptibility to cancers. A number of studies have focused on GSTP1 polymorphism and the risk of various kinds of cancers, including osteosarcoma, ovarian carcinoma, prostate cancer, and bladder cancer,6,8,22,23 but the polymorphism was not considered to be linked with susceptibility to the chemotherapy of osteosarcoma patients. This study was performed to investigate the association between GSTP1 polymorphism and chemotherapy of osteosarcoma.
To the best of our knowledge, our study is the first meta-analysis assessing the association between GSTP1 polymorphism and chemotherapy of osteosarcoma. In this study, an obvious association among tumor response, events, and survival times was found in GSTP1 313A>G(rs1695) under the recessive model, but there was no obvious association between survival times and chemotherapy of osteosarcoma in GSTP1 I105V. We concluded that the GSTP1 313A>G(rs1695) polymorphism may be associated with chemotherapy of osteosarcoma. Although we use this comprehensive analysis to demonstrate the association between GSTP1 polymorphism and chemotherapy of osteosarcoma, some limitations still exist. On the one hand, not all of the studies used controls that were population-based, and some risk factors such as age, family history, and environmental factors were unadjusted estimates, which may cause confounding bias. And on the other hand, the number of studies and the number of samples included in this meta-analysis were relatively small. Therefore, we need additional studies with larger sample size and providing more detailed information to assess the effect of GSTP1 313A>G(rs1695) and I105V polymorphism on osteosarcoma with chemotherapy. In spite of the above shortcomings, our study still had the advantage that a meta-analysis of the association of GSTP1 polymorphism on chemotherapy of osteosarcoma is statistically more powerful than any single study, and we found that I105V gene polymorphism did not show an obvious association with chemotherapy of osteosarcoma. Whether GSTP1 polymorphism can be used as an indicator of the risk of osteosarcoma in patients and as a prognostic marker in osteosarcoma patients undergoing chemotherapy warrants further study.
Acknowledgment
This research was supported by the National Natural Science Foundation of China (grant number 81372004).
Disclosure
The authors report no conflicts of interest in this work.
References
Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. | ||
Fuchs B, Zhang K, Schabel A, Bolander ME, Sarkar G. Identification of twenty-two candidate markers for human osteogenic sarcoma. Gene. 2001;278(1–2):245–252. | ||
Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treat Rev. 2006;32(6):423–436. | ||
Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–790. | ||
Foster L, Dall GF, Reid R, Wallace WH, Porter DE. Twentieth-century survival from osteosarcoma in childhood. Trends from 1933 to 2004. J Bone Joint Surg Br. 2007;89(9):1234–1238. | ||
Caronia D, Patiño-Garcia A, Peréz-Martínez A, et al. Effect of ABCB1 and ABCC3 polymorphisms on osteosarcoma survival after chemotherapy: a pharmacogenetic study. PLoS One. 2011;6(10):e26091. | ||
Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600. | ||
Hengstler JG, Kett A, Arand M, et al. Glutathione S-transferase T1 and M1 gene defects in ovarian carcinoma. Cancer Lett. 1998;130(1–2):43–48. | ||
Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009;19(3):279–282. | ||
Miyake T, Nakayama T, Kagara N, et al. Association of GSTP1 methylation with aggressive phenotype in ER-positive breast cancer. Anticancer Res. 2013;33(12):5617–5623. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Li JZ, Tian ZQ, Jiang SN, Feng T. Effect of variation of ABCB1 and GSTP1 on osteosarcoma survival after chemotherapy. Genet Mol Res. 2014;13(2):3186–3192. | ||
Windsor RE, Strauss SJ, Kallis C, Wood NE, Whelan JS. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: a pilot study. Cancer. 2012;118(7):1856–1867. | ||
Liu S, Yi Z, Ling M, et al. Predictive potential of ABCB1, ABCC3, and GSTP1 gene polymorphisms on osteosarcoma survival after chemotherapy. Tumour Biol. 2014;35(10):9897–9904. | ||
Teng JW, Yang ZM, Li J, Xu B. Predictive role of glutathione S-transferases (GSTs) on the prognosis of osteosarcoma patients treated with chemotherapy. Pak J Med Sci. 2013;29(5):1182–1186. | ||
Yang LM, Li XH, Bao CF. Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac J Cancer Prev. 2012;13(11):5883–5886. | ||
Zhang SL, Mao NF, Sun JY, et al. Predictive potential of glutathione S-transferase polymorphisms for prognosis of osteosarcoma patients on chemotherapy. Asian Pac J Cancer Prev. 2012;13(6):2705–2709. | ||
Hu X, Benson PJ, Srivastava SK, et al. Glutathione S-transferases of female A/J mouse liver and forestomach and their differential induction by anti-carcinogenic organosulfides from garlic. Arch Biochem Biophys. 1996;336(2):199–214. | ||
Autrup H. Genetic polymorphisms in human xenobiotica metabolizing enzymes as susceptibility factors in toxic response. Mutat Res. 2000;464(1):65–76. | ||
Yu Z, Li Z, Cai B, et al. Association between the GSTP1 Ile105Val polymorphism and prostate cancer risk: a systematic review and meta-analysis. Tumour Biol. 2013;34(3):1855–1863. | ||
Wang Z, Xue L, Chong T, Li H, Chen H. Quantitative assessment of the association between glutathione S-transferase P1 Ile105Val polymorphism and bladder cancer risk. Tumour Biol. 2013;34(3):1651–1657. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






