Back to Journals » OncoTargets and Therapy » Volume 9
Association between cyclin D1 G870A polymorphism and hepatocellular carcinoma risk: a meta-analysis
Authors Luo T , Chen J, Liu J, Li H, You X, Wang H, Zhu S, Li L
Received 18 March 2016
Accepted for publication 24 May 2016
Published 21 July 2016 Volume 2016:9 Pages 4483—4489
DOI https://doi.org/10.2147/OTT.S108754
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Tao Luo,1,* Jie Chen,1,* Jun-Jie Liu,2,* Hang Li,2 Xue-Mei You,1 Hong-Liang Wang,1 Shao-Liang Zhu,1 Le-Qun Li1
1Department of Hepatobiliary Surgery, 2Department of Ultrasound, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, People’s Republic of China
*These authors contributed equally to this work
Background: Cyclin D1 (CCND1) G870A polymorphism may be associated with hepatocellular carcinoma (HCC) risk, but the results of previous studies were inconsistent. Available evidence was meta-analyzed to assess their potential association.
Methods: Databases PubMed, EMBASE, Cochrane Library, Web of Science, Chinese Biomedical Literature database, China National Knowledge Infrastructure, and Google Scholar were systematically searched. Meta-analyses were performed to investigate the association of G870A polymorphism with HCC risk by calculating odds ratios (ORs) and 95% confidence intervals (CIs) from the data of relevant case–control studies.
Results: Results of this meta-analysis of six case–control studies involving 1,030 cases and 1,683 controls indicate that G870A polymorphism was not associated with HCC risk in any of the five genetic models tested (recessive model: AA vs GG + AG: OR =1.38, 95% CI =0.95–2.00, P=0.09; dominant model: AG + AA vs GG: OR =1.38, 95% CI =0.87–2.20, P=0.17; homozygous model: AA vs GG: OR =1.60, 95% CI =0.87–2.94, P=0.13; heterozygous model: AG vs GG: OR =1.24, 95% CI =0.86–1.79, P=0.25; allelic model: A vs G: OR =1.30, 95% CI =0.95–1.80, P=0.10). Subgroup analyses according to ethnicity showing marginally significant association between this single nucleotide polymorphism and HCC risk indicate that G870A may be significantly associated with HCC risk in Caucasian populations (recessive model: AA vs GG + AG: OR =2.34, 95% CI =1.60–3.42, P<0.0001; dominant model: AG + AA vs GG: OR =2.44, 95% CI =1.19–4.97, P=0.01; homozygous model: AA vs GG: OR =3.42, 95% CI =1.80–6.50, P=0.0002; allelic model: A vs G: OR =2.06, 95% CI =1.31–3.24, P=0.002), but not in Asian populations.
Conclusion: Available evidence suggests that no significant association between G870A polymorphism and HCC risk was found in either total populations or Asian populations. However, significant association was found in Caucasian populations. These results should be verified and extended in further detailed and well-designed studies involving larger, multiethnic samples.
Keywords: cyclin D1 G870A, single nucleotide polymorphism, hepatocellular carcinoma, meta-analysis
Introduction
Hepatocellular carcinoma (HCC) is considered as the fifth most common neoplasm worldwide, with an increasing incidence and a low survival. It is also the second leading cause of cancer death in males and the sixth leading cause of cancer death in females around the globe.1,2 Highly aggressive and poor prognosis in terrible mortality are the predominant characteristics of HCC throughout the world. Therefore, HCC is a serious fatal disease and has caused serious damages to human health. As a complex and heterogeneous process, the pathogenesis of HCC is not completely understood. Previous studies have established that the development of HCC is linked to interactions of various risk factors, such as chronic infection with hepatitis B virus (HBV) or hepatitis C virus, liver cirrhosis, chronic alcohol use, and exposure to aflatoxin.3–5 Furthermore, recent studies, which highlighted the relationship between gene polymorphisms and the development and progression of HCC, have proposed that genetic factors may contribute to carcinogenesis.6,7
Cyclin D1 (CCND1) is a major regulatory protein that plays a specific role in the cell cycle, promoting the G1 to S phase transition of the cell cycle by binding and activating cyclin-dependent kinase 4 (CDK4) or cyclin-dependent kinase 6 (CDK6).8,9 Overexpression or disordered regulation of CCND1 is a driving force in certain cancers of humans, such as non-small-cell lung cancer10 and breast cancer.11 Due to the eventfulness of CCND1 in the cell division, CCND1 gene single nucleotide polymorphism (SNP) has been investigated in various cancers. G870A (rs603965) is one of the most frequent SNPs in the CCND1 gene, located at codon 242 in the splice donor region of exon 4.12
Numerous case–control studies have investigated a potential association between CCND1 G870A polymorphism and risk of carcinomas. Most of these studies have demonstrated that G870A may be associated with decreased risk of carcinomas, while some failed to detect any significant association.13–19 Similarly, the available case–control studies investigating the possible association between G870A and HCC risk have given inconsistent results.20,21 Related meta-analyses have attempted to synthesize the available evidence on these associations.22,23 Unfortunately, the results of these studies were inconsistent due to small sample sizes involved. Thus, we conducted a meta-analysis involving relevant literature up to date with a larger sample size to provide more comprehensive and reliable insights into investigating the possible associations between G870A polymorphism and HCC risk.
Methods
Search strategy
The databases PubMed, EMBASE, Cochrane Library, Web of Science, Chinese Biomedical Literature database, China National Knowledge Infrastructure, and Google Scholar were systematically searched up to March 15, 2016, to identify all clinical and experimental case–control studies examining associations between G870A polymorphism and HCC risk, using the following search strategy: “Cyclin D1”, “CCND1”, “G870A”, or “rs603965”; “hepatocellular carcinoma”, “liver cancer”, or “HCC”; and “polymorphism”, “polymorphisms”, “SNP”, “variant”, “variation”, “mutation”, “genotype”, or “genetic”. Reference lists in identified articles and reviews were also searched manually to identify additional eligible studies.
Inclusion criteria
Studies included in the meta-analysis were required to meet the following criteria: 1) should have assessed the association of G870A polymorphism with HCC risk in humans, 2) should have had case–control design, 3) should be available as full text, 4) should have contained sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI), including genotype frequencies, and 5) should have been published in English or Chinese language. If multiple publications reported data for the same study sample, only the study with the largest sample size was included.
Data extraction
Two authors (Tao Luo and Shao-Liang Zhu) independently reviewed and extracted the following data from all the included studies: first author’s family name, year of publication, country or district, ethnicity, genotyping methods, total numbers of cases and controls, genotype frequencies in cases and controls, and the P-value associated with Hardy–Weinberg equilibrium (HWE) in controls. Discrepancies were resolved by discussion until consensus was achieved.
Statistical analysis
All statistical tests for this meta-analysis were performed using Review Manager 5.3 (Cochrane Collaboration, Oxford, UK) and Stata 12.0 (StataCorp LP, College Station, TX, USA).
The association between G870A polymorphism and HCC risk was estimated for each polymorphism in different comparison models, including recessive model (AA vs GG + AG), dominant model (AG + AA vs GG), homozygote model (AA vs GG), heterozygote model (AG vs GG), and allelic model (A vs G) in cases and controls by calculating unadjusted ORs with CIs. Significance of pooled ORs was determined using the Z-test, and the P-value <0.05 was considered significant. Meta-analysis was conducted using a fixed-effect model (the Mantel–Haenszel method) when P>0.10 for the Q-test, which indicates lack of heterogeneity among the included studies;24 otherwise, a random-effect model (the DerSimonian–Laird method) was used.25 To explore the sources of heterogeneity among studies, we performed subgroup analyses according to ethnicity. HWE in the control group was assessed using Fisher’s exact test with P<0.05 considered significant.
Publication bias was assessed using Begg’s funnel plot and Egger’s weighted regression, with P<0.05 considered significant.
Results
Description of included studies
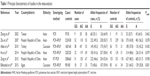
Figure 1 is a flowchart illustrating the search and study selection criteria. A total of 936 potentially relevant articles published up to March 15, 2016, were systematically identified in database searches. Of these, we excluded 923 studies based on the review of titles and abstracts. An additional seven studies were excluded because they were reviews (n=3), meta-analyses (n=3), or did not provide usable data (n=1). As a result, six studies reporting the relationship between G870A polymorphism and HCC risk were included in this meta-analysis involving five English20,21,26–28 and one Chinese29 articles. Thus, a total of six divided case–control design comparisons involving 1,030 cases and 1,683 controls were finally included in the meta-analysis. Of all the eligible studies, four (including 780 HCC cases and 1,343 controls) were conducted in Asian populations,21,26,27,29 and two (including 250 HCC cases and 340 controls) were in Caucasian populations.20,28 The principal characteristics of the recruited studies are presented in Table 1. The distribution of genotypes in controls was consistent with HWE (P>0.05) in all but one study.28
  | Figure 1 Flowchart of study selection for meta-analysis. |
Results of meta-analysis
The main meta-analysis results of the association between G870A polymorphism and HCC risk are shown in Table 2.
  | Table 2 Meta-analysis of the association of CCND1 G870A polymorphism with HCC risk |
Pooled data from the entire population of 1,030 cases and 1,683 controls showed no significant association between G870A polymorphism and HCC risk in any of the five genetic models tested (recessive model: AA vs GG + AG: OR =1.38, 95% CI =0.95–2.00, P=0.09; dominant model: AG + AA vs GG: OR =1.38, 95% CI =0.87–2.20, P=0.17; homozygous model: AA vs GG: OR =1.60, 95% CI =0.87–2.94, P=0.13; heterozygous model: AG vs GG: OR =1.24, 95% CI =0.86–1.79, P=0.25; allelic model: A vs G: OR =1.30, 95% CI =0.95–1.80, P=0.10; Figure 2).
However, subgroup analyses according to ethnicity, marginally significant association between G870A polymorphism, and HCC risk indicate that G870A may be significantly associated with HCC risk in Caucasian populations (recessive model: AA vs GG + AG: OR =2.34, 95% CI =1.60–3.42, P<0.0001; dominant model: AG + AA vs GG: OR =2.44, 95% CI =1.19–4.97, P=0.01; homozygous model: AA vs GG: OR =3.42, 95% CI =1.80–6.50, P=0.0002; allelic model: A vs G: OR =2.06, 95% CI =1.31–3.24, P=0.002), but not in Asian populations (recessive model: AA vs GG + AG: OR =1.07, 95% CI =0.80–1.43, P=0.66; dominant model: AG + AA vs GG: OR =1.02, 95% CI =0.70–1.48, P=0.93; homozygous model: AA vs GG: OR =1.07, 95% CI =0.63–1.81, P=0.81; heterozygous model: AG vs GG: OR =0.99, 95% CI =0.74–1.32, P=0.95; allelic model: A vs G: OR =1.04, 95% CI =0.82–1.31, P=0.77).
Sensitivity analysis
Sensitivity analysis of the G870A meta-analysis was performed by excluding a single individual study each time to reflect the influence to the overall ORs. Excluding the study by Zhu et al29 led to a statistically significant result in the recessive model (AA vs GG + AG: OR =1.57, 95% CI =1.11–2.24, P=0.01), dominant model (AG + AA vs GG: OR =1.64, 95% CI =1.07–2.52, P=0.02), homozygote model (AA vs GG: OR =2.01, 95% CI =1.16–3.49, P=0.01), heterozygote model (AG vs GG: OR =1.42, 95% CI =1.01–1.99, P=0.04), and allele model (A vs G: OR =1.45, 95% CI =1.06–1.99, P=0.02). Therefore, our meta-analysis results should be interpreted with caution.
Publication bias
The risk of publication bias in this meta-analysis was assessed by generating Begg’s funnel plots and performing Egger’s test. Applying the allelic model (A vs G) to data from the total populations, we observed no obvious asymmetry in Begg’s funnel plots (Figure 3A and B) and obtained nonsignificant P-values in Egger’s test for the SNP (allele model: G vs A: t=1.15, P=0.314).
Discussion
CCND1 appears to play an important role in numerous types of cancers, where it modulates the key process during G1 to S phase transition of the cell cycle by binding and activating CDK4 or CDK6. Many studies have investigated G870A SNP in CCND1 and its potential associations with HCC risk and progression, but the results are conflicting. Therefore, we meta-analyzed the available literature as possible as we can to provide more precise estimation of the association.
The study by Elkhashab et al28 demonstrated that SNP of G870A among the Egyptian population was associated with increased risk of HCC. The results of Akkiz et al20 indicated that the G870A polymorphism might be the genetic susceptibility that influences hepatocellular carcinogenesis within the Turkish population studied. Hu et al21 suggested that the G870A polymorphism might play a role in the development of HCC among HBsAg-positive individuals in the Chinese population. Zhu et al29 deduced that G870A was concerned with HCC risk only in females. However, the study by Zeng et al27 found that the G870A genotype had no association with the susceptibility of HBV-related HCC in the Chinese cohort. Zhang et al26 speculated that CCND1 genotype frequencies were similar in cases and controls and were not associated with susceptibility to the development of HCC. Two related meta-analyses by Pabalan et al30 and Chen et al23 drew a semblable conclusion that the G870A polymorphism conferred susceptibility to the development of cancers or digestive tract cancers. Zhao et al22 revealed a significant relationship between the G870A polymorphism and HCC risk in the overall populations. Nevertheless, the meta-analysis by Zhao et al22 contained a study31 investigating the CCND1 polymorphism and onset of hepatoblastoma, instead of HCC, which might lead to an imprecision of the results.
As far as we know, this article is a meta-analysis of the comprehensive assessment that focuses only on the relationship between G870A polymorphism and HCC risk. Compared with those previous meta-analyses, the current one contained three more case–control studies involving 533 cases and 1,268 controls. The current work contained a total of six case–control studies involving 1,030 cases and 1,683 controls and showed that there was no significant association between G870A polymorphism and HCC risk in the overall populations in the five genetic models. The results are consistent with most case–control studies.21,27,29 It seems that G870A polymorphism is not likely to affect HCC risk. However, after being stratified by ethnicity, G870A polymorphism presented as a risk factor for HCC in Caucasian populations but not among Asians in any of the five genetic models. These results were consistent with a related study by Chen et al,23 which suggested that G870A polymorphism may be associated with the risk of digestive tract cancers, specifically among Caucasians. The reason may be that the environment they lived in and the genetic backgrounds influence the relationship between G870A polymorphism and HCC risk. Moreover, the same polymorphisms playing different roles in tumor susceptibility among different ethnic populations have been detected in numerous studies in recent years.32–34
To the best of our knowledge, the findings of this meta-analysis provide more comprehensive and detailed analysis of whether G870A polymorphism is correlated with HCC risk. Nevertheless, the results are limited by the designs of the included studies. First, the controls in the included studies were not uniformly defined, such that it was impossible to determine which controls were population based and which were hospital based. Second, the P-value of HWE was <0.05 in the study by Elkhashab et al,28 suggesting that this study population was not representative of the broader target population. Nevertheless, we decided to retain this study in the meta-analysis because excluding it did not significantly affect the pooled ORs (data not shown). Third, HCC risk may also depend on numerous confounding factors that we did not account for in the meta-analysis, such as HBV status, tumor status, sex, and age. We could not adjust for these factors using subgroup analyses because most studies either did not report these baseline data or aggregated them in different ways. Fourth, methods used to test for polymorphisms were not uniform and they varied in sensitivity and specificity, which may reduce the robustness of the meta-analysis. Finally, this meta-analysis included only two studies from Caucasian populations and none from African; larger, well-designed studies in Caucasian and African populations are warranted to reevaluate these associations.
Conclusion
Available evidence suggests that no significant association between G870A polymorphism and HCC risk was found in either total populations or Asian populations. However, significant association was found in Caucasian populations. These results should be verified and extended in further detailed and well-designed studies involving larger, multiethnic samples.
Acknowledgments
This research was supported by the National Science and Technology Major Special Project (2012ZX10002010001009), the Self-Raised Scientific Research Fund of the Ministry of Health of Guangxi Province (Z2015621 and Z2014241), and the Guangxi University of Science and Technology Research Fund (KY2015LX056).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. | ||
Severi T, van Malenstein H, Verslype C, van Pelt JF. Tumor initiation and progression in hepatocellular carcinoma: risk factors, classification, and therapeutic targets. Acta Pharmacol Sin. 2010;31(11):1409–1420. | ||
Liu Y, Pan S, Liu L, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7(4):e35145. | ||
Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology. 2010;51(6):1972–1978. | ||
Mao CF, Qian WY, Wu JZ, Sun DW, Tang JH. Association between the XRCC3 Thr241Met polymorphism and breast cancer risk: an updated meta-analysis of 36 case-control studies. Asian Pac J Cancer Prev. 2014;15(16):6613–6618. | ||
Shin A, Lee KM, Ahn B, et al. Genotype-phenotype relationship between DNA repair gene genetic polymorphisms and DNA repair capacity. Asian Pac J Cancer Prev. 2008;9(3):501–505. | ||
Bienvenu F, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J Biol Chem. 2001;276(20):16840–16847. | ||
Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88(3):405–415. | ||
Myong NH. Cyclin D1 overexpression, p16 loss, and pRb inactivation play a key role in pulmonary carcinogenesis and have a prognostic implication for the long-term survival in non-small cell lung carcinoma patients. Cancer Res Treat. 2008;40(2):45–52. | ||
Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–4367. | ||
Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. | ||
Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, Mittal B. Role of BCL2 (ala43thr), CCND1 (G870A) and FAS (A-670G) polymorphisms in modulating the risk of developing esophageal cancer. Cancer Detect Prev. 2007;31(3):225–232. | ||
Matthias C, Branigan K, Jahnke V, et al. Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998;4(10):2411–2418. | ||
Wang SS, Cozen W, Severson RK, et al. Cyclin D1 splice variant and risk for non-Hodgkin lymphoma. Hum Genet. 2006;120(2):297–300. | ||
Yu CP, Yu JC, Sun CA, Tzao C, Ho JY, Yen AM. Tumor susceptibility and prognosis of breast cancer associated with the G870A polymorphism of CCND1. Breast Cancer Res Treat. 2008;107(1):95–102. | ||
Buch S, Zhu B, Davis AG, et al. Association of polymorphisms in the cyclin D1 and XPD genes and susceptibility to cancers of the upper aero-digestive tract. Mol Carcinog. 2005;42(4):222–228. | ||
Grieu F, Malaney S, Ward R, Joseph D, Iacopetta B. Lack of association between CCND1 G870A polymorphism and the risk of breast and colorectal cancers. Anticancer Res. 2003;23(5b):4257–4259. | ||
Han J, Colditz GA, Hunter DJ. Lack of associations of selected variants in genes involved in cell cycle and apoptosis with skin cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15(3):592–593. | ||
Akkiz H, Bayram S, Bekar A, Akgöllü E, Ozdil B. Cyclin D1 G870A polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: case-control study. Cancer Epidemiol. 2010;34(3):298–302. | ||
Hu Z, Zhou Z, Xiong G, et al. Cyclin D1 G870A polymorphism and the risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35(6):5607–5612. | ||
Zhao Y, He HR, Wang MY, et al. Cyclin D1 G870A gene polymorphism and risk of leukemia and hepatocellular carcinoma: a meta-analysis. Genet Mol Res. 2015;14(2):5171–5180. | ||
Chen B, Cao L, Yang P, Zhou Y, Wu XT. Cyclin D1 (CCND1) G870A gene polymorphism is an ethnicity-dependent risk factor for digestive tract cancers: a meta-analysis comprising 20,271 subjects. Cancer Epidemiol. 2012;36(2):106–115. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Zhang YJ, Chen SY, Chen CJ, Santella RM. Polymorphisms in cyclin D1 gene and hepatocellular carcinoma. Mol Carcinog. 2002;33(2):125–129. | ||
Zeng Z, Tu J, Cheng J, et al. Influence of CCND1 G870A polymorphism on the risk of HBV-related HCC and cyclin D1 splicing variant expression in Chinese population. Tumour Biol. 2015;36(9):6891–6900. | ||
Elkhashab MN, Refaey MM, Radwan MI, et al. Association between Cyclin D1 G870A polymorphism and the risk of hepatocellular carcinoma among Egyptian population. Afro Egypt J Infect Endem Dis. 2015;5(4):297–304. | ||
Zhu ZZ, Cong WM, Xian ZH, et al. Correlation of Cyclin D1 polymorphism with genetic susceptibility to hepatocellular carcinoma. Chin J Cancer Prev Treat. 2007;14(20):1521–1523. | ||
Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2773–2781. | ||
Pakakasama S, Chen TT, Frawley W, et al. CCND1 polymorphism and age of onset of hepatoblastoma. Oncogene. 2004;23(27):4789–4792. | ||
Zhang L, Jiang Y, Wu Q, et al. Gene-environment interactions on the risk of esophageal cancer among Asian populations with the G48A polymorphism in the alcohol dehydrogenase-2 gene: a meta-analysis. Tumour Biol. 2014;35(5):4705–4717. | ||
Chen B, Zhou Y, Yang P, Liu L, Qin XP, Wu XT. CDH1 −160C>A gene polymorphism is an ethnicity-dependent risk factor for gastric cancer. Cytokine. 2011;55(2):266–273. | ||
Ju W, Kim JW, Park NH, et al. Matrix metalloproteinase-1 promoter polymorphism and epithelial ovarian cancer: does ethnicity matter? J Obstet Gynaecol Res. 2007;33(2):155–160. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.