Back to Journals » Infection and Drug Resistance » Volume 13
Assessment and Comparison of Two Serological Approaches for the Surveillance of Health Workers Exposed to SARS-CoV-2
Authors Russo A , Calò F, Di Fraia A, Starace M , Minichini C, Gentile V, Angelillo IF , Coppola N
Received 18 September 2020
Accepted for publication 18 November 2020
Published 17 December 2020 Volume 2020:13 Pages 4501—4507
DOI https://doi.org/10.2147/IDR.S282652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Antonio Russo,1 Federica Calò,1 Alessandra Di Fraia,1 Mario Starace,1 Carmine Minichini,1 Valeria Gentile,1 Italo Francesco Angelillo,2 Nicola Coppola1 On behalf of the Vanvitelli-COVID-19 Group
1Department of Mental Health and Public Medicine, Section of Infectious Diseases, University of Campania, Naples, Italy; 2Department of Experimental Medicine, University of Campania, Naples, Italy
Correspondence: Nicola Coppola Department of Mental Health and Public Medicine, Section of Infectious Diseases
University of Campania, via: L. Armanni 5, Naples 80131, Italy
Tel +39 0815666719
Fax +39 0815666013
Email [email protected]
Background and Aim: The aim of the present study was to assess the diagnostic performance of an LFA compared with an ELISA test in a cohort of HWs operating in a COVID-19 unit of a teaching hospital in southern Italy.
Methods: We performed an observational, prospective, interventional study including 65 COVID-19 unit personnel. On a total of 196 serum samples (at least 2 serum samples for each HW), LFA and ELISA tests for SARS-COV-2 IgG and IgM were performed. Also, 32 serum samples of SARS-CoV-2 RNA positive patients at least 21 days before sampling, and 30 serum samples of patients obtained up to November 2019, before COVID-19 outbreak in China, were used as positive and negative controls, respectively.
Findings: Of the 65 HWs enrolled, 6 were positive in LFA; overall, of the 196 serum samples, 20 were positive in LFA. All ELISA tests performed on serum samples collected from HWs were negative. The specificity of LFAs was 90.77% considering the 65 HWs and 89.80% considering all the 196 health workers serum samples analyzed. Considering the data on HWs, ELISA test for SARS-COV-2 antibodies showed a specificity of 100%, including all the 196 serum samples collected, and 100% including the 65 HWs. The ELISA and LFAs performed after 21 days last COVID-19 patient was discharged were all negative.
Conclusion: LFAs compared to ELISA tests result in less specificity, considering COVID-19 negative personnel and patients. Thus, LFAs seem to be not adequate in the active surveillance of HWs.
Keywords: health workers, COVID-19, SARS-CoV-2, anti-IgG SAR-COV-2, active surveillance
Introduction
From the first description of pneumonia of unknown cause detected in Wuhan, China, at the end of December 2019, the pandemic caused by the novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is dramatically accelerating worldwide.1
Given the significant impact of Coronavirus disease (COVID)-19 outbreak, it is of fundamental importance to implement the use of diagnostic tests for the quick identification of suspected cases. This might facilitate contact tracing of people exposed to the virus in order to put in place infection control measures, necessary to reduce the further spread of the pandemic.
Health workers (HWs) are at high risk of infection while caring for COVID-19 patients and they can be responsible for nosocomial transmission clusters; thus, the surveillance of HWs is a key point in the management of SARS-CoV-2 infection. Clinical passive surveillance with self-assessment for fever or other suggestive symptoms for COVID-19 is recommended for all exposed HWs. However, this strategy may lack information from asymptomatic or mildly symptomatic subjects. For this reason, it is of utmost importance that all HWs undergo, in addition to clinical passive surveillance, microbiological active surveillance with nucleic acid amplification tests (NAATs) or serological tests.
Although NAATs in respiratory samples represent the gold standard for the diagnosis of current infection with SARS-CoV-2, they have some limitations, such as the cost, the medium turnaround time ranged between 4 and 24 hours2 and the fact that they require specialized personnel; in addition, insufficient viral RNA at the point of detection may lead to false-negative results.3 Numerous serological immunoassays have been introduced for the identification of immunity against SARS-CoV-2; they could be easily implemented in any hospital with a much wider application than molecular tests.4 The IgM-IgG antibody test exhibited a useful adjunct to RT-PCR detection, and improved the accuracy in COVID-19 diagnosis.5 Compared to NAATs, antibody assays are often faster, less expensive, require no trained technicians to operate4 and the requirement for specimen quality is less stringent than for RNA-based assays.4 Their sensitivity may not depend on the type of serological test used only, but also on the timing of disease presentation and exposure to the virus; in fact, detection of IgM antibodies may take 3 to 6 days to appear and IgG antibodies typically appear after 8 days,6–8 features that make them unreliable in the early stages of the illness. Point-of-care lateral flow assays (LFAs) automated chemiluminescence immunoassay (CLIA) and enzyme-linked immunosorbent assay (ELISA) are the most widely used commercial tests. Rapid IgM/IgG tests by LFAs can be efficiently used in the surveillance of HWs since they are inexpensive and simple tests giving results in 10–15 minutes, thus particularly suitable to population scale diagnosis of SARS-CoV-2 in both high-income countries and low- and middle-income countries.9
The aim of the present study was to assess the diagnostic performance of two serological assays, in particular of an LFAs compared with an ELISA test in a cohort of HWs in a COVID-19 unit of a teaching hospital in southern Italy.
Methods
Study Design
We performed an observational, prospective study in the COVID-19 unit of the University of Campania “L. Vanvitelli”, Naples: the unit included 24 beds for the care of COVID-19 patients and was active from 22 March to 3 July 2020; the nursing staff included one nurse for every four beds.
All HWs employed at this unit were included in the study. They all followed the infection prevention and control (IPC) procedures according to the Azienda Ospedaliera Universitaria Vanvitelli protocol (prot.287/2020). According to these procedures, passive and active surveillance were performed for all HWs: passive surveillance was based on body temperature control at the beginning and at the end of every work shift and on COVID-19-related-symptoms check; active surveillance was done by a lateral flow assay (LFA) for SARS-CoV-2 IgG and IgM every 7 days. In case of fever (body temperature>37.5°C) and/or COVID-19-related symptoms and/or positive LFA, a NAAT for SARS-COV-2-RNA on nasopharyngeal swab was performed.
To satisfy the aims of the study, from May, 2 2020 to June, 8 2020, apart from the LFAs, surveillance was implemented with ELISA tests for SARS-COV-2 IgG and IgM on at least 2 serum samples for each HW and a further serum sample was collected 21 days since the last COVID-19 patient was discharged, as shown in Figure 1. A NAAT for SARS-CoV-2 on nasopharyngeal swabs for each HW was performed in May 2020 (Figure 1). Moreover, in August 2020, at least 45 days after the discharge of the last patient, tests for SARS-COV-2 IgG and IgM by ELISA and a NAAT for SARS-CoV-2 on nasopharyngeal swabs were performed in all HWs.
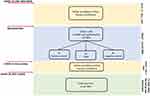 |
Figure 1 Study design. |
In order to assess the sensibility and specificity of LFA and ELISA, as positive controls, we tested 32 serum samples of COVID-19 patients, samples collected 21–32 days from the first NAAT positivity, and, as negative controls, 30 serum samples of patients obtained from July 2019 up to November 2019 (8 in July, 6 in August, 7 in September, 7 in October, 2 in November 2019). Moreover, all the 30 subjects did not have a history of traveling abroad in the last month before blood sampling.
The study was approved by the Ethics Committee of the University of Campania, Naples (n°10877/2020). All procedures performed in this study were in accordance with the ethics standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethics standards. Informed consent was obtained from all participants included in the study.
Lateral Flow Assays (LFAs)
As LFAs, the Xiamen Wiz Biotec (Xiamen, China) Diagnostic Kit (Colloidal Gold) for IgM/IgG antibodies for SARS-CoV-2 was used according to the manufacturer’s instructions: if the control line did not appear the test was considered invalid; in the presence of the control line, any visible band for IgG or IgM was indicative of a positive result.10
ELISA for SARS-CoV-2 Antibodies
For the ELISA test, we used WANTAI SARS-CoV-2 Ab ELISA (Xiamen, China), that tests for total antibodies to the SARS-CoV-2 receptor binding domain. The test was used according to the manufacturer’s instructions on at least 2 sera for each HW during the activity of the COVID-19 unit and one serum at the end: if the specimen had absorbance less than the cut-off value (calculated using the negative control absorbance and a standard value), it was considered negative; otherwise, if it had absorbance more than the cut-off value, it was considered positive; the specimens with cut-off ratio absorbance between 0.9 and 1.1 were considered borderline and were re-tested. The specificity and sensitivity declared was respectively 100% and 94.5%.
NAAT for SARS-CoV-2
On all nasopharyngeal swabs collected for the study, SARS-CoV-2 RNA was sought by real-time polymerase chain reaction (PCR). Viral RNA was extracted from 140 µl of nasopharyngeal swab using a microspin column (QIAamp RNA viral kit, Qiagen GmbH, Hilden, Germany) in Qiagen platform. SARS-CoV-2 RNA was identified by performing a real-time PCR in a Light cycler 480 (Roche Diagnostics, Branchburg, NJ, USA) by REALQUALITY RQ-2019-nCoV (Ab ANALITICA, Padova, Italy), following the manufacturer’s instructions. The PCR primers targeted SARS-CoV-2-gene RDRP and gene E.
Statistical Analysis
Continuous variables were expressed as median (IQR); categorical variables were expressed as number (%). Confidence intervals of sensitivity and specificity are “exact” Clopper-Pearson confidence intervals. All analyses were performed using STATA v16.
Results
Assessment of Sensitivity and Specificity by Positive and Negative Controls
As positive controls, a total of 32 serum samples from hospitalized patients with a diagnosis of COVID-19 were tested by LFAs and ELISA. All sera but 3 resulted positive in LFAs; all sera but one resulted positive in ELISA (Tables 1 and 2). In particular, one serum resulted negative by both LFAs and ELISA, and 2 sera only in LFAs. Thus, the sensitivity of LFAs was 91.4% (95% CI: 76.9–98.2), that of ELISA test was 97.0% (95% CI: 84.2–99.9) (Tables 1 and 2).
 |
Table 1 Result of LFAs in Different Settings |
 |
Table 2 Results of ELISA Tests in Different Settings |
As negative controls, 30 serum samples collected before the COVID-19 outbreak were tested by LFA and ELISA. All serum samples resulted negative, both for LFAs and ELISA, showing a specificity for both tests of 100% (95% CI: 88.4–100.0) (Tables 1 and 2).
The Characteristics of the Health Workers Enrolled
Sixty-five HWs agreed to be included in the study between May 2 and June 8. Of the HWs enrolled, 25 (38.5%) were male, the median age was 32 (IQR 28–43), 5 were patient-care technicians, 35 were nurses, 25 physicians (14 infectious diseases, 5 cardiologists and 6 pulmonologists) (Table 3). During the study no HW enrolled had fever or symptoms compatible with COVID-19. No-one showed any deviation from the protocol or close contact with SARS-COV-2 infected patients without personal protective equipment.
 |
Table 3 Description of HWs Enrolled |
Following our protocol, between May 8–13, between July 15–21 and between 17–30 August all HWs performed NAATs and no-one resulted positive for SARS-COV-2 infection.
From July 15 to 21 and from August 17 to 30, the ELISA test was performed in all 65 HWs enrolled and resulted negative in all.
Assessment of Sensitivity and Specificity by Analysis of HWs
Overall, the 65 HWs enrolled performed 196 LFAs and ELISA tests for SARS-COV-2 antibodies: LFA and ELISA were performed three times in 64 HWs and four times in 1 subject.
The median between the first and the last LFA and ELISA tests, excluding one performed after 21 days after the last COVID-19 patient was discharged, was 16 days (range 7–37).
Of the 65 HWs enrolled, 6 (9.23%) tested positive in LFA; similarly, of the 196 LFA tests performed, 20 (10.2%) resulted in positive (Table 1). Of the 6 positive subjects, 5 tested positive in all LFAs performed and one only tested positive in 1 test. All ELISA tests performed on serum samples collected from HWs were negative (Table 3).
Considering the data on the HWs, the ELISA test for SARS-COV-2 antibodies showed a specificity of 100% (95% CI: 98.1–100.0), including all 196 serum samples collected, and 100% (95% CI: 94.48–100.00) including the 65 HWs (Table 2). The specificity of LFAs was 90.77% (95% CI: 80.98–96.54) considering only the 65 subjects and 89.80% (95% CI: 84.68–93.65) considering all 196 serum samples analyzed (Table 1).
Including the data of the negative controls on our HWs and the test performed 21 days after the last COVID-19 patient was discharged, the specificity of LFAs was 90.65% (95% CI: 74.98–98.02) and the specificity of the ELISA test was 100% (95% CI: 98.7–100.0) (Tables 1 and 2).
Discussion
Although the management of the HWs working in COVID-19 units is fundamental in the infection control strategy, it lacks information for the best surveillance management.
There is no validated screening program for HWs involved in COVID-19 management, and further studies are required to identify the best active surveillance. The WHO published interim guidance on “Risk assessment and management of exposure of health workers in the context of COVID-19”, defining high and low risk depending on exposure.11 HWs exposed to a high-risk infection should stop all healthcare interaction with patients and get tested for COVID-19, should stay in quarantine and daily self-monitor temperature and respiratory symptoms for 14 days after the last day of exposure to a COVID-19 patient. HWs considered at a low risk should self-monitor temperature and respiratory symptoms daily for 14 days after the last day of exposure to a COVID-19 patient.11
However, various strategies of screening HWs have been put in place including active and passive surveillance.12,13 Considering active surveillance, various strategies have been used, based on NAAT or serological surveillance or both in order to reduce the limits of each test. Widely is the low sensitivity of serological tests in the early phase of infection,2,12–17 due to the natural antibody dynamic with 100% positivity for IgG after 17–19 days from infection.18 This delay can be limiting in a setting of HWs constantly exposed to SARS-CoV-2, hospital outbreaks. NAAT is one of the most expensive tests on the market, with the need for expert staff and adequate facilities, which limit its use in screening, making the serological test a preference especially in some population settings.
The present study assessed two serological approaches, a rapid serological and ELISA test, for the surveillance of HWs exposed to SARS-CoV-2. To our knowledge, this study is the first study that compared the diagnostic performance of LFAs with ELISA tests in a large cohort of HWs in a COVID-19 unit. In our cohort of HWs the specificity of LFAs was lower than that identified for the ELISA test (89.80% vs 100%); including also the patients admitted to our hospital before the outbreak in China, the specificity was 89.30% for LFAs and 100% for the ELISA test. Moreover, considering the patients hospitalized in our COVID-unit, the sensitivity was lower in LFAs compared with that identified for ELISA (90.65% vs 96.88%). The low specificity of LFAs leaves a margin of doubt regarding the infection until the results of NAAT, creating difficulties in work organization and also psychological and social problems. The limitation of LFAs was suggested also in other settings.19,20 For example, in an emergency room department of a tertiary hospital in northern Italy on 110 subjects (30 healthy volunteers, 30 COVID-19-positive patients and 50 patients at their first access to an emergency room department with fever and respiratory syndrome), Cassaniti et al found that the sensitivity of LFA was 18.4%, specificity 91.7%, while NPV was 26.2% and PPV was 87.5%.17 Ong et al, including patients presenting at a teaching hospital with respiratory symptoms, showed a very heterogeneous sensitivity using 5 different LFA kits, ranging from 2/20 (10%) to 11/20 (55%); after choosing the most sensitive LFA they tested 228 patients and showed a sensitivity of 43% and a specificity of 98%.2
Although there was a variation in performance characteristics, antibody testing with LFAs offers an additional value for estimating infection rate among HWs in resource-limited countries where PCR or ELISA are not readily available. Indeed, the main benefit of LFAs is the availability of real-time results and the advantage of being easily repeatable making them potentially useful in hospital setting for HWs surveillance.
The present study has, however, some limits to consider. First, this study included only one of the LFA and ELISA tests available on market. Second, the number of positive controls are limited, but in line with the literature.2,14 Third, fortunately, no HW was infected during the COVID-19 experience, but this means in our analysis we cannot compare the sensitivity of tests during the early phase of infection.
In conclusion, considering the data of the present study and those of the studies available in the literature, the LFA test does not seem to be indicated in the active surveillance of HWs.
Abbreviations
LFA, lateral flow assay; NAAT, nucleic acid amplification test; HWs, Health workers; ELISA, enzyme-linked immunosorbent assay.
Acknowledgments
We thank all the Vanvitelli COVID-19 group (Nicola Coppola, Caterina Sagnelli, Stefania De Pascalis, Maria Stanzione, Gianfranca Stornaiuolo, Angela Cascone, Salvatore Martini, Margherita Macera, Caterina Monari, Federica Calò, Carmine Minichini, Mario Starace, Alessandra di Fraia, Andrea Bianco, Antonio Russo, Valeria Gentile, Clarissa Camaioni, Giulia De Angelis, Giulia Marino, Roberta Astorri, Ilario De Sio, Marco Niosi, Serena Borrelli, Vincenzo Carfora, Benito Celia, Maria Ceparano, Salvatore Cirillo, Maria De Luca, Marco Di Mauro, Grazia Mazzeo, Marco Migliaccio, Filiberto Fausto Mottola, Giorgio Paoli, Riccardo Ricciolino, Giorgio Spiniello, Nicoletta Verde) for the help in the preparation and critical revision of the paper.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
“POR Campania FESR 2014-2020-Avviso per l’acquisizione di manifestazioni di interesse per la realizzazione di servizi di ricerca e sviluppo per la lotta contro il Covid-19” (DGR n. 140 del 17 marzo 2020), Project: “IDENTIFICAZIONE DEI FATTORI DEMOGRAFICI, CLINICI, VIROLOGICI, GENETICI, IMMUNOLOGICI E SIEROLOGICI ASSOCIATI AD OUTCOME SFAVOREVOLE NEI SOGGETTI CON COVID-19”, Regione Campania, Italy.
Disclosure
All the authors of the manuscript declare that they have no conflict of interest in connection with this paper.
References
1. WHO coronavirus disease (COVID-19) Dashboard | WHO coronavirus disease (COVID-19) dashboard. Available from: https://covid19.who.int/.
2. Ong DSY, de Man SJ, Lindeboom FA, Koeleman JGM. Comparison of diagnostic accuracies of rapid serological tests and ELISA to molecular diagnostics in patients with suspected coronavirus disease 2019 presenting to the hospital. Clin Microbiol Infect. 2020;26(8):
3. Russo A, Minichini C, Starace M, Astorri R, Calò F, Coppola N. Current status of laboratory diagnosis for covid-19: a narrative review. Infect Drug Resist. 2020;13:2657–2665. doi:10.2147/IDR.S264020
4. Xiao SY, Wu Y, Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J Med Virol. 2020;92(5):464–467. doi:10.1002/jmv.25702
5. Liu R, Liu X, Yuan L, et al. Analysis of adjunctive serological detection to nucleic acid test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection diagnosis. Int Immunopharmacol. 2020;86:106746. doi:10.1016/j.intimp.2020.106746
6. Zhang M, Jiang Y, Xiao X, Peng M, Peng F, Gong G. Differences in IP-10, TLR4 and IRF5/3 between SVR and non-SVR HCV-1 patients treated with PEG-IFN and ribavirin. Mol Med Rep. 2017;15(4):2318–2324. doi:10.3892/mmr.2017.6229
7. Xiang F, Wang X, He X, et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis. 2020. doi:10.1093/cid/ciaa461
8. Lee HK, Lee BH, Seok SH, et al. Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J Vet Sci. 2010;11(1):165–167. doi:10.4142/jvs.2010.11.2.165
9. Grant BD, Anderson CE, Williford JR, et al. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal Chem. 2020;92(16):11305–11309. doi:10.1021/acs.analchem.0c01975
10. Wantai SARS-CoV-2 Rapid test. Beijing Wantai Biological Pharmacy Enterprise C o., Ltd. Beijing, China. Available from: http://www.dbaitalia.it/newsletter/wantai_sars-cov-2_ab_rapid_test_-_ce_ifu.pdf.
11. Macera M, De Angelis G, Sagnelli C, Coppola N. Clinical presentation of covid-19: case series and review of the literature. Int J Environ Res Public Health. 2020;17(14):1–11. doi:10.3390/ijerph17145062
12. Fusco FM, Pisaturo M, Iodice V, et al. COVID-19 among healthcare workers in a specialist infectious diseases setting in Naples, Southern Italy: results of a cross-sectional surveillance study. J Hosp Infect. 2020;105(4):596–600. doi:10.1016/j.jhin.2020.06.021
13. Lombardi A, Consonni D, Carugno M, et al. Characteristics of 1573 healthcare workers who underwent nasopharyngeal swab testing for SARS-CoV-2 in Milan, Lombardy, Italy. Clin Microbiol Infect. 2020;26(10):
14. Döhla M, Boesecke C, Schulte B, et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health. 2020;182:170–172. doi:10.1016/j.puhe.2020.04.009
15. Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128. doi:10.1016/j.jcv.2020.104413
16. Van Elslande J, Houben E, Depypere M, et al. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26(8):1082–1087. doi:10.1016/j.cmi.2020.05.023
17. Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020;92(10):1724–1727. doi:10.1002/jmv.25800
18. Butner JL, Gupta N, Fabian C, et al. Onsite treatment of HCV infection with direct acting antivirals within an opioid treatment program. J Subst Abuse Treat. 2017;75:49–53. doi:10.1016/j.jsat.2016.12.014
19. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi:10.1038/s41591-020-0897-1
20. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. doi:10.1093/cid/ciaa310
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
