Back to Journals » The Application of Clinical Genetics » Volume 14
Appropriate Clinical Genetic Testing of Hemochromatosis Type 2–4, Including Ferroportin Disease
Authors Kowdley DS, Kowdley KV
Received 13 May 2021
Accepted for publication 18 July 2021
Published 12 August 2021 Volume 2021:14 Pages 353—361
DOI https://doi.org/10.2147/TACG.S269622
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Martin Maurer
Devan S Kowdley, Kris V Kowdley
Liver Institute Northwest and Elson S. Floyd College of Medicine, Washington State University, Seattle, WA, USA
Correspondence: Kris V Kowdley
Liver Institute Northwest, 3216 NE 45th Place Suite 212, Seattle, WA, 98105, USA
Tel +1 206-536-3030
Fax +1 206-524-7429
Email [email protected]
Abstract: Hereditary hemochromatosis (HH) is an inherited iron overload disorder due to a deficiency of hepcidin, or a failure of hepcidin to degrade ferroportin. The most common form of HH, Type 1 HH, is most commonly due to a homozygous C282Y mutation in HFE and is relatively well understood in significance and action; however, other rare forms of HH (Types 2– 4) exist and are more difficult to identify and diagnose in clinical practice. In this review, we describe the clinical characteristics of HH Type 2– 4 and the mutation patterns that have been described in these conditions. We also review the different methods for genetic testing available in clinical practice and a pragmatic approach to the patient with suspected non-HFE HH.
Keywords: non-HFE, hemochromatosis, juvenile hemochromatosis, ferroportin disease, transferrin-receptor 2, HAMP, hepcidin
Introduction
Iron is an essential metal for its biological role in hemoglobin and myoglobin to deliver oxygen to tissues, and as a cofactor for a host of enzymes.1,2 Body iron stores are regulated at the level of intestinal absorption, and there is no physiologic mechanism other than blood loss via menses for elimination of excess iron.3 Therefore, prevention of iron overload must occur at the level of absorption in the duodenum.
Hepcidin
Hepcidin is a 25-amino acid peptide that regulates systemic iron homeostasis by inhibition of iron absorption.4 Hepcidin originates in the liver in response to elevated iron levels and binds to ferroportin (FPN) on intestinal cells, resulting in a reduction of cellular iron export.5 Hepcidin has major effects on control of iron absorption in enterocytes and macrophages, and is crucial to the regulation of overall serum iron levels.6
Iron Overload Disorders
Iron overload disorders may be classified as primary or secondary iron overload disorders. Primary iron overload disorders have a genetic basis, and can be attributed to either low hepcidin production or decreased binding interactions between hepcidin and FPN, the transmembrane cellular iron exporter.7,8 These primary iron overload disorders are classified as different types of hereditary hemochromatosis. These disorders have variable phenotypic expression but all share the central defect of decreased hepcidin activity due to different mutations. The most frequent form of hereditary hemochromatosis is one of the most common genetic disorders among Caucasians, with a homozygote frequency of approximately 1 in 250 individuals of Northern European descent.9,10 Type 1 or classical hereditary hemochromatosis, is due to mutations in HFE, the gene encoding the HFE protein.11 Patients with HFE-associated HH may be asymptomatic; chronic fatigue and arthropathy may be early signs.3 The most common HFE mutation is a guanine to alanine substitution at position 845 of the HFE gene, resulting in a cysteine to tyrosine change (p.C282Y).11 This mutation is inherited in an autosomal recessive pattern, and p.C282Y heterozygotes generally do not express the hemochromatosis phenotype.12 Clinical penetrance of p.C282Y hereditary hemochromatosis may be as low as 2%, and is influenced by other factors leading to iron overload such as excess alcohol intake or chronic hepatitis C13. The mutation responsible for p.C282Y was originally identified in 1996 by a full physical mapping of the 3-Mb genomic region around the HLA-A3 allele on chromosome 6 in diagnosed HH patients.11 Along with p.C282Y, a second missense mutation in HFE was also identified in the same physical mapping, with a cytosine to guanine change resulting in a substitution from histidine to aspartic acid, p.H63D.11 p.H63D heterozygotes may have an increased likelihood of iron overload if they are compound heterozygotes with the p.C282Y mutation; this form of HH is known as type 1B hereditary hemochromatosis. However, heterozygosity or homozygosity for H63D alone is unlikely to result in expression of the hemochromatosis phenotype.13
The p.C282Y mutation is present in a large proportion of patients who present with the phenotype of hereditary hemochromatosis (80.6%).14 Testing for HFE-associated hereditary hemochromatosis is readily available, as a multiplex-PCR can be performed to identify p.H63D and p.C282Y concurrently.15
Other more rare forms of hereditary hemochromatosis relevant to the clinician may be caused by mutations in hemojuvelin (Type 2A HH), hepcidin antimicrobial peptide (Type 2B HH), transferrin receptor 2 (Type 3A HH), or ferroportin (Type 4 HH) (Table 1).11,16–18 A recent publication by Sandhu et al is a comprehensive collection of cases of identified non-HFE hereditary hemochromatosis19. It is important to note that iron overload may also be caused by inherited mutations in the H-ferritin gene (Type 5 HH); however, Type 5 HH has only been identified in one Japanese family.20 Additionally, aceruloplasminemia can be the source of iron overload and present with increased serum ferritin, but is not generally considered a form of hereditary hemochromatosis.21
 |
Table 1 Hereditary Hemochromatosis Subtypes, Genes Affected and Most Common Mutations |
Molecular Testing (General)
Molecular diagnosis of monogenic inherited diseases is based on identifying variants that may explain phenotypic patterns.23 It is estimated that approximately 8% of all live births will be diagnosed with a genetic abnormality, with the majority due to monogenic autosomal variants.24 Linkage mapping of recessive traits using restriction fragment length polymorphisms (RFLP) was first described as a method of diagnosis for recessive traits within consanguineous relatives in 1987,25 and is particularly useful for autosomal recessive monogenic diseases such as cystic fibrosis.26 However, linkage analysis limits resolution and may be unable to identify candidate genes.23 In the late 20th century, identification efforts for pathogenic genes were typically carried out to identify monogenic diseases, and typically within single pedigrees.27
The Human Genome Project (HGP) represented a watershed moment in sequencing and identification of genetic disease.28 The HGP identified 19,000 protein-coding (exome) genes, representing approximately 1% of the genome; however, a majority of phenotypes and pathogenic variants occur in exomic loci.29,30 Massively parallel sequencing (MPS) or next-generation sequencing (NGS) drastically increased the data-to-price ratio of sequencing exomes, reducing genes of interest from 3 million base pairs of the genome to 20,000 single-nucleotide polymorphisms (SNPs) of exomes.31 There are two approaches to exome sequencing: whole-exome sequencing and array-based sequencing.32 NGS can be used as a diagnostic tool for rare diseases and can reveal novel mutations within known clinical genes of interest.33 Whole-exome sequencing (WES) focuses screening on exomes of genes of interest, as interpretation of introns is limited.34 This method has proved effective in postmortem diagnosis in early sudden death due to monogenic disease variants.35 Exome sequencing is valuable for diagnosis of prenatal disease, and WES of parent and child can be used to identify de novo mutations of interest.31 For example, WES has expanded genetic diagnosis of terminal prenatal conditions after ultrasound abnormalities, even when karyotyping appears normal.34 Definitive diagnosis after WES has been estimated to be approximately 25 to 35% and is especially useful in determining autosomal dominant mutations.36 Chromosomal microarray analysis (CMA) examines changes in genes within a known chromosomal region.37 CMA has higher sensitivity for nucleotide additions and deletions and is typically used to determine copy number variants (CNVs) or SNPs of clinical significance.38 Genetic testing for diagnosis of disease is a rapidly evolving field, with genotyping not only being used for diagnosis of diseases but may also to guide individualized treatment regimens.39
Hereditary hemochromatosis may also be identified through molecular testing but is not as straightforward as simply testing for p.C282Y. In a genotype analysis of African Americans, primary iron overload was found to be due to varying causes and not as straightforward as the relative frequency of p.C282Y in Caucasian patients with primary iron overload.40 Since the clinical consequences and diagnosis of HH have been described in detail, in this review we discuss and elucidate features and diagnosis of non-HFE hemochromatosis. A variety of options exist for clinicians to test for non-HFE hemochromatosis (Table 2). Genetic testing for HH is usually ordered in patients with elevated serum transferrin-iron saturation and/or ferritin; the diagnosis of non-HFE hemochromatosis is usually only suspected in patients who test negative for the p.C282Y mutation.41
 |
Table 2 Available Testing for Non-HFE Related HH Subtypes, as of 6/22/21 |
Type 2A- HJV
Type 2 hereditary hemochromatosis, also known as juvenile hemochromatosis (JH), is an early-onset form of HH. Two subtypes of juvenile hemochromatosis have been identified: type 2A and type 2B. Type 2A hereditary hemochromatosis is caused by mutations in HJV, located on chromosome 1q which encodes hemojuvelin (HJV), which is expressed in the same tissues as hepcidin.48 The median age of presentation for Type 2A is 25 years, and while Type 1 hemochromatosis shows a male predominance, HH Type 2A affects sexes equally.2 Type 2A HH has a distinct set of clinical symptoms compared to Type 1 HH, and characteristically includes iron overload, hypogonadotropic hypogonadism, diabetes and most significantly, cardiomyopathy.49 Type 2A HH may also result in amenorrhea in women, further increasing body iron stores.50 If untreated, cardiomyopathy is a frequent cause of death among JH probands.51,52 Based on family members of one lineage, it was suggested that clinical penetrance of HJV mutations is higher than mutations in HFE.50 The earliest age at which patients present with symptoms of iron overload associated with HJV mutations was age 4 in an African American patient with the homozygous nonsense mutation p.R54X.53
Originally known as HFE2, HJV was first mapped by Papanikolaou et al, from a Greek family.16 Six candidate mutations were identified based on polymorphisms compared to controls in ten Greek families, one Canadian family and one French family in juvenile hemochromatosis patients without p.C282Y mutations.16 The most common mutation in HJV is a guanine to thymine transition originally discovered by Sanger sequencing, resulting in a glycine to valine substitution in position 320 of HJV (p.G320V).16 WES also revealed a homozygous substitution in HJV from guanine to thymine in position 959, confirming WES validity in diagnosis of p.G320V hemochromatosis.54 The second mutation in HJV identified by Papanikolaou et al was a substitution of thymine to adenine at position 665, known as p.I222N.16 The p.I281T mutation has been detected both as a homozygous mutation in a Greek patient and as compound heterozygous with the nonsense mutation p.C321X in Chinese patients.16,55 Mutations in HJV are not always identified in patients with juvenile hemochromatosis; HJV mutations in Japan (p.D249H, p.Q312X) have resulted in HH phenotypes with onset in middle age.56
In a PCR-RFLP study in Alabama of Caucasians and African Americans there were no p.G320V homozygotes and only one heterozygote for p.I222N was found, indicating the relative rarity of these mutations in the population.57 It has been proposed that WES should be considered for a possible diagnosis of Type 2A HH only after demonstration of elevated ferritin and transferrin saturation, and negative HFE and p.G320V testing.54 Classical symptoms of Type 2A HH accompanied by secondary hypothyroidism and iron overload in the pituitary may be due to a frameshift mutation from a cytosine deletion at position 697 of the HJV gene, resulting in a stop codon at amino acid 245 of HJV.58 Commercially available testing for Type 2A HH includes single gene panels for p.G320V available from Fulgent43 and Invitae;44 exome sequencing available from Blueprint Genetics,42 Prevention Genetics,45 the Valencian Institute of Microbiology (Ivami)46 and Invitae.44 Companies identified for testing were identified by the use of search engine, using keywords “non-HFE hemochromatosis testing, HJV testing”.
Type 2B- HAMP
The HAMP gene located on chromosome 19 directly encodes HAMP, a propeptide that is cleaved into the 25 amino acid hepcidin. Clinical features of HAMP hemochromatosis are similar to those of Type 2A HH.13 HAMP mutations are relatively rare and heterozygosity in HAMP can be observed as a modifier of the p.C282Y HFE phenotype.59 Microsatellite analysis of chromosome 19q13 in two individuals of Italian and Greek descent with juvenile hemochromatosis (JH) but with no HJV mutations revealed two HAMP mutations: a frameshift mutation resulting from a guanine deletion at position 3 of HAMP (93delG) and a cytosine to guanine transversion resulting in a nonsense mutation (p.R56X).60 This led HAMP-associated HH to be classified as a subtype of JH. PCR screening of the HAMP gene in 21 unrelated JH patients identified a missense from cysteine to arginine (p.C70R), removing a stabilizing disulfide in the structure of hepcidin.61 Mutations in HAMP have also been observed in both Russia and Portugal, and the overall geographic range is not completely defined.61,62
A mutation in the 5ʹUTR of HAMP mRNA has been identified in Portuguese patients, creating a mutant initiation codon resulting in iron overload phenotypes from mistranslation of hepcidin.63
As in Type 2A HH, testing for Type 2B HH includes exome sequencing available from Blueprint Genetics42 Prevention Genetics,45 Valencian Institute of Microbiology (Ivami)46 and Invitae,44 and next-generation sequencing from Fulgent.43 Companies identified for testing were identified by the use of search engine, using keywords “non-HFE hemochromatosis testing, HAMP testing”.
Type 3- TFR2
Type 3 hereditary hemochromatosis is caused by mutations in the transferrin receptor 2 (TFR2) gene on chromosome 7; TFR2 is relatively long with a length of 20 kB and many mutations and polymorphisms have been identified.64 It was initially thought that HFE formed dimers in the duodenum with transferrin receptor 2 (TFR2), but TFR2 acts as an independent regulator of hepcidin in hepatocytes.65 TFR2-associated hemochromatosis is described as intermediate in clinical presentation between HFE-HH and HJV-HH, and mutations in TFR2 can produce a form of HH resembling juvenile hemochromatosis.66,67
TFR2-associated hereditary hemochromatosis is rare, with approximately 65 cases in 44 family lineages across a wide geographic area.68 Although TFR2-HH was initially discovered in Italy, subsequent cases were found in Scotland, Spain, Japan and Taiwan.68–71 However, TFR2-associated HH is still most commonly found in Italy and Japan.
Mutations in TFR2 were first discovered upon analysis of two unrelated families of Sicilian origin (one heavily inbred) presenting with hemochromatosis but with no mutations in p.C282Y.17 Linkage mapping excluding HFE and HJV revealed homozygosity in the region of TFR2 on chromosome 7; microsatellite analysis confirmed a cytosine to guanine substitution in exon 6 of TFR2, creating a nonsense mutation (p.Y250X) in TFR2.17 The second mutation in TFR2 to be discovered was within a consanguineous family, and was an insertion of a cytosine into exon 2, resulting in a nonsense mutation at amino acid 60 (p.E60X).72
A homozygous deletion in exon 16 of TFR2 resulting in deletion of amino acids 594–597 was found in a proband of Northern Italian descent with known iron overload, and intrafamilial haplotyping of affected siblings detected early-onset HH phenotype.69 Three potential mutations of TFR2 were proposed in a Scottish man in 2006, including p.R396X, and p.G792R, but could not be directly attributed to the observed HH phenotype.70 Despite the relative rarity of HH in Asian populations, WES of TFR2 in a Taiwanese woman found a mutation of guanosine to adenosine in exon 11 (p.R481H) resulting in extreme iron overload and diabetes mellitus.71 The first described change in TFR2 mRNA levels was a splice site mutation resulting in skipping of exon 4 in TFR2 reported in a woman of Southern Italian descent.73 In a study of four unrelated Spanish patients with HH symptoms and no mutations in HFE, HJV, HAMP and SLC40A1, homozygous mutations in TFR2 were discovered, including a missense mutation p.G792R, and nonsense mutations p.Q306X and p.Q672X.68
TFR2 sequencing as a single gene is available from Invitae44 and Fulgent,43 and whole exome sequencing is available from Blueprint Genetics,42 Ivami46 and Prevention Genetics.45 Additionally, Baylor Genetics Laboratories has created a sequence-specific test designed to detect private mutations.47 Companies identified for testing were identified by the use of search engine, using keywords “non-HFE hemochromatosis testing, TFR2 testing”.
Type 4A- FPN Disease
Type 4A hereditary hemochromatosis is also known as classic ferroportin disease (FD), and can be categorized as its own disease associated with iron overload. Outside of HFE-HH, FD is the most prevalent form of genetic iron overload regardless of race.18,74 FD is caused by pathogenic mutations in Fpn or SLC40A1, which encodes ferroportin (FPN), also known as Solute Carrier Family 40 Member 1 protein. FD is distinct from hereditary hemochromatosis because it is not associated with high transferrin-iron saturation or low hepcidin concentrations, and unlike other forms of HH, FD is typically inherited in an autosomal dominant pattern.74,75 In FD, the hepcidin-FPN interaction marking FPN for degradation is disturbed, resulting in a heterogeneity of clinical symptoms.22
The most common FD genotypes are an alanine to aspartic acid at residue 77 (p.A77D) substitution in exon 3 of the FPN gene and a deletion of valine at residue 162 (p.V162del).8,22 p.A77D was detected by PCR amplification of exons in the FPN gene comparing probands and control groups.8 Two missense mutations causing FD (p.I152F and p.L233P) were identified in probands by denaturing high-performance liquid chromatography (DHPLC), where Fpn mutations were transfected in vitro into kidney cell lines and then injected into zebrafish embryos, using immunofluorescence to detect FPN localization and degradation in the cell.76
Molecular testing for SLC40A1 includes whole exome sequencing is available from Blueprint Genetics,42 Ivami46 and Prevention Genetics,45 and next-generation sequencing from Fulgent.43 Companies identified for testing were identified by the use of search engine, using keywords “non-HFE hemochromatosis testing, FPN testing”.
Type 4B
Type 4B hereditary hemochromatosis also results from a mutation in SLC40A1; however, Type 4B HH is not caused by a change in expression of SLC40A1, but rather is a gain-of-function mutation in FPN. This “resistant” form of FPN is no longer susceptible to degradation by hepcidin binding and remains hyperactive on the cell membrane.77 Types 4A and 4B HH can be distinguished clinically by the finding of a low to normal transferrin saturation (TS) in type 4A and high TS in type 4B.22 MRI has been suggested as a non-invasive tool to distinguish between the gain-of-function and loss-of-function forms of Type 4 HH.78
The first form of Type 4B HH was discovered by RFLP analysis of Thai and Vietnamese patients by an autosomal dominant mutation p.C326Y creating a resistant FPN with no effect on protein localization.79 The cysteine residue at position 326 is involved in a disulfide interaction between hepcidin and FPN, and any change in the cysteine at position 326 of FPN results in minimal hepcidin internalization.80,81 An Australian proband with cirrhosis and parenchymal iron overload onset was identified with relatively early onset of disease at 32 years old, with a change in the asparagine residue to aspartic acid at position 144.82 A splice mutation in SLC40A1 was detected by PCR analysis of HFE, HJV, HAMP, TFR2 compared against a cDNA library in a middle-aged Chinese woman with unexplained iron overload.83
Conclusion
Primary iron overload not due to mutations in HFE can be due to a variety of mutations in HJV, HAMP, TFR2 and/or FPN. These rare forms of non-HFE HH and difficult to diagnose with certainty, given the variety of mutations, many of which are private. Moreover, compound heterozygosity can add challenges to the diagnosis when HFE mutations accompany HJV or HAMP mutations (Figure 1). It is also important to note that the majority of suspected HH cases are not attributable to any genetic cause, so adequate clinical suspicion is necessary for diagnosis of genetic HH and it is important to eliminate secondary causes of iron overload. Clinicians should always seek alternative explanations for iron overload in patients with iron overload in the absence of HFE mutations before considering a diagnosis of non-HFE hereditary hemochromatosis given the rarity of these disorders. Patient genetic history and ancestry should be taken into account before diagnosis and treatment with therapeutic phlebotomy.13 Genetic testing for non-HFE HH should be considered in patients with documented iron overload after other causes if iron overload have been excluded, especially among patients with a family history of iron overload. Several clinical laboratories offer genetic testing for these forms of HH and appropriate use of genetic testing may provide confirmation of the diagnosis and prognosis.
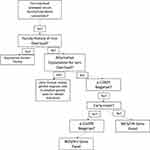 |
Figure 1 Flowchart for clinician approach to patients with unexplained iron overload. |
Disclosure
Dr Kris V Kowdley reports grants, personal fees from La Jolla, personal fees from Protagonist, during the conduct of the study. The authors report no other conflicts of interest in this work.
References
1. Wang C-Y, Babitt JL. Liver iron sensing and body iron homeostasis. Blood. 2019;133:18–29.
2. Gerhard GS, Paynton BV, DiStefano JK. Identification of genes for hereditary hemochromatosis. Methods Mol Biol. 2018;1706:353–365.
3. Brissot P, Pietrangelo A, Adams PC, et al. Haemochromatosis. Nat Rev Dis Primers. 2018;4(1):18016. doi:10.1038/nrdp.2018.16
4. Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276(11):7806–7810. doi:10.1074/jbc.M008922200
5. Anderson GJ, Frazer DM. Current understanding of iron homeostasis. Am J Clin Nutr. 2017;106(Supplement 6):1559S–1566S. doi:10.3945/ajcn.117.155804
6. Kowdley KV, Gochanour EM, Sundaram V, Shah RA, Handa P. Hepcidin signaling in health and disease: ironing out the details. Hepatol Commun. 2021;5(5):723–735. doi:10.1002/hep4.1717
7. Siddique A, Kowdley KV. Review article: the iron overload syndromes. Aliment Pharmacol Ther. 2012;35(8):876–893. doi:10.1111/j.1365-2036.2012.05051.x
8. Montosi G, Donovan A, Totaro A, et al. Autosomal-dominant hemochrom-atosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108(4):619–623. doi:10.1172/JCI200113468
9. Adams PC, Reboussin DM, Barton JC, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352(17):1769–1778. doi:10.1056/NEJMoa041534
10. Gochee PA, Powell LW. What’s new in hemochromatosis. Curr Opin Hematol. 2001;8(2):98–104. doi:10.1097/00062752-200103000-00007
11. Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I–like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. doi:10.1038/ng0896-399
12. Zaloumis SG, Allen KJ, Bertalli NA, et al. Natural history of HFE simple heterozygosity for C282Y and H63D: a prospective 12-year study. J Gastroenterol Hepatol. 2015;30(4):719–725. doi:10.1111/jgh.12804
13. Kowdley KV, Brown KE, Ahn J, Sundaram V. ACG clinical guideline: hereditary hemochromatosis. Am J Gastroenterol. 2019;114(8):1202–1218. doi:10.14309/ajg.0000000000000315
14. European Association For The Study Of The Liver. EASL clinical practice guidelines for HFE hemochromatosis. J Hepatol. 2010;53:3–22.
15. Arts HH, Eng B, Waye JS. Multiplex allele-specific PCR for simultaneous detection of H63D and C282Y HFE mutations in hereditary hemochromatosis. J Appl Lab Med. 2018;3(1):10–17. doi:10.1373/jalm.2017.024984
16. Papanikolaou G, Samuels ME, Ludwig EH, et al. Mutations in HFE2 cause iron overload in chromosome 1q–linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. doi:10.1038/ng1274
17. Camaschella C, Roetto A, Calì A, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25(1):14–15. doi:10.1038/75534
18. Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32(1):131–138. doi:10.1016/j.bcmd.2003.08.003
19. Sandhu K, Flintoff K, Chatfield MD, et al. Phenotypic analysis of hemochromatosis subtypes reveals variations in severity of iron overload and clinical disease. Blood. 2018 Jul 5;132(1):101–110. doi:10.1182/blood-2018-02-830562
20. Kato J, Fujikawa K, Kanda M, et al. A mutation, in the iron-responsive element of H ferritin mRNA, causing autosomal dominant iron overload. Am J Hum Genet. 2001;69(1):191–197. doi:10.1086/321261
21. Roberti MDRF, Borges Filho HM, Gonçalves CH, Lima FL. Aceruloplasminemia: a rare disease - diagnosis and treatment of two cases. Rev Bras Hematol Hemoter. 2011;33(5):389–392. doi:10.5581/1516-8484.20110104
22. Mayr R, Janecke AR, Schranz M, et al. Ferroportin disease: a systematic meta-analysis of clinical and molecular findings. J Hepatol. 2010;53(5):941–949. doi:10.1016/j.jhep.2010.05.016
23. Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(S3):228–237. doi:10.1038/ng1090
24. Baird PA, Anderson TW, Newcombe HB, Lowry RB. Genetic disorders in children and young adults: a population study. Am J Hum Genet. 1988;42:677–693.
25. Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236(4808):1567–1570. doi:10.1126/science.2884728
26. Kerem B, Rommens J, Buchanan J, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245(4922):1073–1080. doi:10.1126/science.2570460
27. Claussnitzer M, Cho JH, Collins R, et al. A brief history of human disease genetics. Nature. 2020;577(7789):179–189. doi:10.1038/s41586-019-1879-7
28. Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
29. Chong JX, Buckingham K, Jhangiani S, et al. The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97(2):199–215. doi:10.1016/j.ajhg.2015.06.009
30. Makrythanasis P, Antonarakis SE. Pathogenic variants in non-protein-coding sequences. Clin Genet. 2013;84(5):422–428. doi:10.1111/cge.12272
31. Stranneheim H, Wedell A. Exome and genome sequencing: a revolution for the discovery and diagnosis of monogenic disorders. J Intern Med. 2016;279(1):3–15. doi:10.1111/joim.12399
32. Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130. doi:10.1038/gim.2017.247
33. Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking mendelian disease using exome sequencing. Genome Biol. 2011;12(9):228. doi:10.1186/gb-2011-12-9-228
34. Jelin AC, Vora N. Whole exome sequencing: applications in prenatal genetics. Obstet Gynecol Clin North Am. 2018;45(1):69–81. doi:10.1016/j.ogc.2017.10.003
35. Salfati EL, Spencer EG, Topol SE, et al. Re-analysis of whole-exome sequencing data uncovers novel diagnostic variants and improves molecular diagnostic yields for sudden death and idiopathic diseases. Genome Med. 2019;11(1):83. doi:10.1186/s13073-019-0702-2
36. Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi:10.1056/NEJMoa1306555
37. Chang Y-S, Lin C-Y, Huang H-Y, Chang J-G, Kuo H-T. Chromosomal microarray and whole-exome sequence analysis in Taiwanese patients with autism spectrum disorder. Mol Genet Genomic Med. 2019;7(12):e996. doi:10.1002/mgg3.996
38. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi:10.1016/j.ajhg.2010.04.006
39. Franceschini N, Frick A, Kopp JB. Genetic testing in clinical settings. Am J Kidney Dis. 2018;72(4):569–581. doi:10.1053/j.ajkd.2018.02.351
40. Barton JC, Acton RT, Rivers CA, et al. Genotypic and phenotypic heterogeneity of African Americans with primary iron overload. Blood Cells Mol Dis. 2003;31(3):310–319. doi:10.1016/S1079-9796(03)00166-9
41. Santos PCJL, Krieger JE, Pereira AC. Molecular diagnostic and pathogenesis of hereditary hemochromatosis. Int J Mol Sci. 2012;13(2):1497–1511. doi:10.3390/ijms13021497
42. Blueprint Genetics. Available from: https://blueprintgenetics.com/tests/panels/metabolic-disorders/hereditary-hemochromatosis-panel/.
43. Fulgent Genetics. Available from: https://www.fulgentgenetics.com/products/disease/raredisease.html.
44. Invitae. Available from: https://www.invitae.com/en/physician/tests/05201/.
45. Prevention Genetics. Available from: https://www.preventiongenetics.com/testInfo?val=Hereditary+Hemochromatosis+Panel.
46. Valencian Institute of Microbiology (Ivami). Available from: https://www.ivami.com/en/genetic-testing-human-gene-mutations-diseases-neoplasias-and-pharmacogenetics/4301-genetic-testing-hereditary-hemochromatosis-i-hamp-hfe-hfe2-slca40a1-i-and-i-tfr2-i.
47. Baylor Genetics. Available from: https://baylorgenetics.com/.
48. Pagani A, Silvestri L, Nai A, Camaschella C. Hemojuvelin N-terminal mutants reach the plasma membrane but do not activate the hepcidin response. Haematologica. 2008;93(10):1466–1472. doi:10.3324/haematol.12508
49. Li C-X, Zhang L, Wang P, Sun L. Clinicopathological diagnosis and treatment of juvenile hemochromatosis. Chin Med J. 2019;132(24):3018–3020. doi:10.1097/CM9.0000000000000547
50. Lee PL, Beutler E, Rao SV, Barton JC. Genetic abnormalities and juvenile hemochromatosis: mutations of the HJV gene encoding hemojuvelin. Blood. 2004;103(12):4669–4671. doi:10.1182/blood-2004-01-0072
51. Cherfane C, Lee P, Guerin L, Brown K. A late presentation of a fatal disease: juvenile hemochromatosis. Case Rep Med. 2013;2013:875093. doi:10.1155/2013/875093
52. Filali M, Le Jeunne C, Durand E, et al. Juvenile hemochromatosis HJV-related revealed by cardiogenic shock. Blood Cells Mol Dis. 2004;33(2):120–124. doi:10.1016/j.bcmd.2004.05.001
53. Murugan RC, Lee PL, Kalavar MR, Barton JC. Early age-of-onset iron overload and homozygosity for the novel hemojuvelin mutation HJV R54X (exon 3; c.160A→T) in an African American male of West Indies descent. Clin Genet. 2008;74(1):88–92. doi:10.1111/j.1399-0004.2008.01017.x
54. Farrell CP, Parker CJ, Phillips JD. Exome sequencing for molecular characterization of non-HFE hereditary hemochromatosis. Blood Cells Mol Dis. 2015;55(2):101–103. doi:10.1016/j.bcmd.2015.04.002
55. Lv T, Zhang W, Xu A, et al. Non- HFE mutations in haemochromatosis in China: combination of heterozygous mutations involving HJV signal peptide variants. J Med Genet. 2018;55(10):650–660. doi:10.1136/jmedgenet-2018-105348
56. Koyama C, Hayashi H, Wakusawa S, et al. Three patients with middle-age-onset hemochromatosis caused by novel mutations in the hemojuvelin gene. J Hepatol. 2005;43(4):740–742. doi:10.1016/j.jhep.2005.06.024
57. Barton JC, Rivers CA, Niyongere S, Bohannon SB, Acton RT. Allele frequencies of hemojuvelin gene (HJV) I222N and G320V missense mutations in white and African American subjects from the general Alabama population. BMC Med Genet. 2004;5(1):29. doi:10.1186/1471-2350-5-29
58. Santiago de Sousa Azulay R, Magalhães M, Tavares MDG, et al. Novel mutation in the Hemojuvelin Gene (HJV) in a patient with juvenile hemochromatosis presenting with insulin-dependent diabetes mellitus, secondary hypothyroidism and hypogonadism. Am J Case Rep. 2020;21:e923108. doi:10.12659/AJCR.923108
59. Jacolot S, Le Gac G, Scotet V, et al. HAMP as a modifier gene that increases the phenotypic expression of the HFE pC282Y homozygous genotype. Blood. 2004;103(7):2835–2840. doi:10.1182/blood-2003-10-3366
60. Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33(1):21–22. doi:10.1038/ng1053
61. Roetto A, Daraio F, Porporato P, et al. Screening hepcidin for mutations in juvenile hemochromatosis: identification of a new mutation (C70R). Blood. 2004;103(6):2407–2409. doi:10.1182/blood-2003-10-3390
62. Potekhina ES, Lavrov AV, Samokhodskaya LM, et al. Unique genetic profile of hereditary hemochromatosis in Russians: high frequency of C282Y mutation in population, but not in patients. Blood Cells Mol Dis. 2005;35(2):182–188. doi:10.1016/j.bcmd.2005.06.012
63. Matthes T, Aguilar-Martinez P, Pizzi-Bosman L, et al. Severe hemochromatosis in a Portuguese family associated with a new mutation in the 5′-UTR of the HAMP gene. Blood. 2004;104(7):2181–2183. doi:10.1182/blood-2004-01-0332
64. Glöckner G, Scherer S, Schattevoy R, et al. Large-scale sequencing of two regions in human chromosome 7q22: analysis of 650 kb of genomic sequence around the EPO and CUTL1 loci reveals 17 genes. Genome Res. 1998;8(10):1060–1073. doi:10.1101/gr.8.10.1060
65. Rishi G, Crampton EM, Wallace DF, Subramaniam VN, Avila M. In situ proximity ligation assays indicate that hemochromatosis proteins Hfe and transferrin receptor 2 (Tfr2) do not interact. PLoS One. 2013;8(10):e77267. doi:10.1371/journal.pone.0077267
66. Radio FC, Majore S, Binni F, et al. TFR2-related hereditary hemochromatosis as a frequent cause of primary iron overload in patients from Central-Southern Italy. Blood Cells Mol Dis. 2014;52(2–3):83–87. doi:10.1016/j.bcmd.2013.08.003
67. Pietrangelo A, Caleffi A, Henrion J, et al. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128(2):470–479. doi:10.1053/j.gastro.2004.11.057
68. Joshi R, Shvartsman M, Morán E, et al. Functional consequences of transferrin receptor-2 mutations causing hereditary hemochromatosis type 3. Mol Genet Genomic Med. 2015;3(3):221–232. doi:10.1002/mgg3.136
69. Girelli D, Bozzini C, Roetto A, et al. Clinical and pathologic findings in hemochromatosis type 3 due to a novel mutation in transferrin receptor 2 gene. Gastroenterology. 2002;122(5):1295–1302. doi:10.1053/gast.2002.32984
70. Lee PL, Barton JC. Hemochromatosis and severe iron overload associated with compound heterozygosity for TFR2 R455Q and two novel mutations TFR2 R396X and G792R. Acta Haematol. 2006;115(1–2):102–105. doi:10.1159/000089474
71. Hsiao P-J, Tsai K-B, Shin S-J, et al. A novel mutation of transferrin receptor 2 in a Taiwanese woman with type 3 hemochromatosis. J Hepatol. 2007;47(2):303–306. doi:10.1016/j.jhep.2007.04.014
72. Roetto A, Totaro A, Piperno A, et al. New mutations inactivating transferrin receptor 2 in hemochromatosis type 3. Blood. 2001;97(9):2555–2560. doi:10.1182/blood.V97.9.2555
73. Pelucchi S, Mariani R, Trombini P, et al. Expression of hepcidin and other iron-related genes in type 3 hemochromatosis due to a novel mutation in transferrin receptor-2. Haematologica. 2009;94(2):276–279. doi:10.3324/haematol.13576
74. Pietrangelo A. Ferroportin disease: pathogenesis, diagnosis and treatment. Haematologica. 2017;102(12):1972–1984. doi:10.3324/haematol.2017.170720
75. Sham RL, Phatak PD, Nemeth E, Ganz T. Hereditary hemochromatosis due to resistance to hepcidin: high hepcidin concentrations in a family with C326S ferroportin mutation. Blood. 2009;114(2):493–494. doi:10.1182/blood-2009-04-216226
76. Girelli D, De Domenico I, Bozzini C, et al. Clinical, pathological, and molecular correlates in ferroportin disease: a study of two novel mutations. J Hepatol. 2008;49(4):664–671. doi:10.1016/j.jhep.2008.05.028
77. Majore S, Bonaccorsi Di Patti MC, Valiante M, et al. Characterization of three novel pathogenic SLC40A1 mutations and genotype/phenotype correlations in 7 Italian families with type 4 hereditary hemochromatosis. Biochim Biophys Acta. 2018;1864(2):464–470. doi:10.1016/j.bbadis.2017.11.006
78. Pietrangelo A, Corradini E, Ferrara F, et al. Magnetic resonance imaging to identify classic and nonclassic forms of ferroportin disease. Blood Cells Mol Dis. 2006;37:192–196. doi:10.1016/j.bcmd.2006.08.007
79. Viprakasit V, Merryweather-Clarke AT, Chinthammitr Y, et al. Molecular diagnosis of the first ferroportin mutation (C326Y) in the far east causing a dominant form of inherited iron overload. Blood. 2004;104(11):3204. doi:10.1182/blood.V104.11.3204.3204
80. Preza GC, Ruchala P, Pinon R, et al. Minihepcidins are rationally designed small peptides that mimic hepcidin activity in mice and may be useful for the treatment of iron overload. J Clin Invest. 2011;121(12):4880–4888. doi:10.1172/JCI57693
81. Fernandes A, Preza GC, Phung Y, et al. The molecular basis of hepcidin-resistant hereditary hemochromatosis. Blood. 2009;114(2):437–443. doi:10.1182/blood-2008-03-146134
82. Wallace DF, Clark RM, Harley HAJ, Subramaniam VN. Autosomal dominant iron overload due to a novel mutation of ferroportin1 associated with parenchymal iron loading and cirrhosis. J Hepatol. 2004;40(4):710–713. doi:10.1016/j.jhep.2003.12.008
83. Zhang W, Lv T, Huang J, Ou X. Type 4B hereditary hemochromatosis associated with a novel mutation in the SLC40A1 gene: a case report and a review of the literature. Medicine. 2017;96:e8064.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
