Back to Journals » OncoTargets and Therapy » Volume 13
Apolipoprotein C1 (APOC1): A Novel Diagnostic and Prognostic Biomarker for Cervical Cancer
Authors Shi X , Wang J, Dai S , Qin L, Zhou J , Chen Y
Received 7 September 2020
Accepted for publication 26 November 2020
Published 15 December 2020 Volume 2020:13 Pages 12881—12891
DOI https://doi.org/10.2147/OTT.S280690
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xiu Shi,1– 3,* Juan Wang,1,* Shouqian Dai,4 Lingyan Qin,5 Jinhua Zhou,1 Youguo Chen1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, People’s Republic of China; 2Clinical Research Center of Obstetrics and Gynecology, Jiangsu Key Laboratory of Clinical Immunology, Soochow University, Suzhou, Jiangsu Province, People’s Republic of China; 3Jiangsu Institute of Clinical Immunology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, People’s Republic of China; 4Department of Emergency Medicine, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, People’s Republic of China; 5Department of Pathology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Youguo Chen; Jinhua Zhou Email [email protected]; [email protected]
Background: Previous reports showed that APOC1 was associated with several cancers but the function of APOC1 in cervical cancer was unknown. This study aimed to investigate the clinical effect and function of APOC1 in cervical cancer.
Materials and Methods: In this study, the relative expression of APOC1 in cervical cancer was detected by RT-qPCR. In order to determine the cell proliferation and migration and invading ability and apoptosis more accurately, we used CCK8 assay, Edu assay, wound healing assay, migration and invasion assay, flow cytometry assay, co-immunoprecipitation, proteomics and Western blot by silencing and overexpressing APOC1, respectively. The role of APOC1 on tumor progression was explored in vitro and vivo.
Results: The relative expression of APOC1 in cervical cancer tissues was up-regulated (P< 0.05). In cervical cancer cell lines, silencing of APOC1 restrained cell progression and EMT, while over-expression of APOC1 accelerated cell progression and EMT in vivo and vitro (P< 0.05).
Conclusion: APOC1 acts as an oncogene in cervical cancers and knockdown of APOC1 inhibited cervical cancer cells growth in vitro and in vivo. There is a close relationship between the relative expression of APOC1 and clinical outcome in cervical cancer patients.
Keywords: APOC1, cervical cancer, progression, prognosis
Introduction
Cervical cancer is considered to be one of the most common gynecological malignant tumors in the world and highly occurred in developing countries,1–5 which is a serious threat to women's health. Globally, cervical cancers rank the fourth one among female tumor causes of death and the incidence of cervical cancer ranks the first one among female genital malignancies in China.6,7 The invasion and metastasis of cervical cancers are the main cause of death in patients and lymphatic metastasis is one of the main poor prognosis of cervical cancers.8–10 Therefore, that we further studied about biological processes of cervical cancer metastasis, and excavated the key regulatory factors and signal transduction network, and developed new therapeutic targets and strategies, and improved the survival rate of patients, has very important clinical significance. However, the mechanism of cervical cancer has not been fully elucidated.
APOC1 is a member of the apolipoprotein family that ACTS not only to transport lipids and stabilize the lipoprotein structure but also to regulate pathological processes including diabetes, Alzheimer’s and inflammation.11–14 In recent years, some studies have found that ApoA1,15,16 ApoB,17–19 ApoE20,21 and other apolipoproteins22 are abnormally expressed in tumors and are related to the prognosis of patients. Therefore, the role of apolipoprotein family in the occurrence and development of tumors has attracted more attention. For example, some studies have shown that M2-type macrophages could deliver ApoE through exosomes to promote gastric cancer cell metastasis.23–25 More recent studies of ApoC1’s relationship with tumors have been limited to clinical observations,26–31 such as ApoC1 elevations, have been reported more frequently about hormone resistance therapy for prostate cancer, prognosis of TRIPLE plus breast cancer, and aggressive progression of lung cancer. ApoC1 has been reported to promote the proliferation of prostate cancer cells and protect against pancreatic cancer cell apoptosis and so on. But the mechanism and signaling pathway of ApoC1’s role in tumors is unclear.
This study shows that APOC1 expression is significantly up-regulated in cervical cancers and APOC1 promoted cervical cancer cell progression and was involved in EMT signal way. It is suggested that APOC1 could act as a promising biomarker and novel therapeutic target for cervical cancers.
Materials and Methods
Tissue Sample
Human cervical cancer tissues and corresponding normal tissues were obtained from the First Affiliated Hospital of Soochow University. The Ethical Committee of the First Affiliated Hospital of Soochow University for Clinical Research approved this study. All the samples were obtained with the patients’ informed consent. No smoking subjects were included in our study. All the patients were diagnosed by two experienced pathologists. The non-tumorous tissue samples were at least 2cm from the edge of the tumor, contained no obvious tumor cells, and were also evaluated by the pathologists.
Cell Line and Culture
The HeLa and SiHa human cervical cancer cells were bought from ATCC and maintained in DMEM and RPMI-1640 culture medium supplemented with 10% of fetal bovine serum (FBS) and incubated at 37 °C in a 5% CO2 incubator.
Transfection
SiRNA were bought from GenePharma (Shanghai, China). si-APOC1-2 sense (5-GAAACACACTGGAGGACAA-3), antisense (5ʹ-CUUUGUGUGACCUCCUGUU-3ʹ); si-APOC1-1sense (5ʹ-GCTGAAGGAGTTTGGAAACAC-3ʹ). Si-NC (Negative Control siRNA) and si-APOC1 were purchased from Gene Pharma. Fubio (Suzhou, China) was the provider of plasmids pcDNA3.1-APOC1 and pcDNA3.1-NC (negative control). Cells in the logarithmic growth were harvested and inoculated in 6-well plates. Cells were cultured for at least 24 hrs before transfection. Cells transfection with the corresponding vector were collected after 48 hrs transfection, the siRNAs and plasmids were transfected into cervical cancer cells using Lipofectamine 3000 (Invitrogen, USA).
RT-PCR
The total RNA was extracted from the specimens and cervical cancers using TRIzol reagent (Invitrogen, USA) based on the product descriptions. APOC1 primers sense: 5ʹ-TCCAGTGCCTTGGATAAGCTG-3ʹ and reverse:5ʹ-GGCTGATGAGTTCCCGAGC-3ʹ GAPDH primers forward: 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ, reverse: 5-’ATCCGTTGACTCCGACCTTCAC-3ʹ.32–35 The cDNA was synthesized from the whole RNA using the Prime Script RT Reagent Kit with gDNA Eraser (Takara, Dalian). SYBR Premix Ex Taq II (Takara, Dalian) was used to detect the APOC1, etc., expression levels by RT-qPCR. The endogenous controls were GAPDH. The relative quantification method (2−ΔΔCt) was used to calculate the expressions that have been normalized to endogenous controls.
CCK-8 Assay
Hela cells and SiHa cells were plated into 96-well plates overnight. On the second day, APOC1 siRNA oligonucleotides and plasmids were transfected according to the lipofectamine3000 operating manual. At 0, 24, 48 and 72 hrs after transfection, replaced the medium with Cell Counting Kit-8 and incubated at 37 °C for 60 mins. Subsequently, the absorbance was measured at 450 nm with a Multiskan GO microplate spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Edu Analysis
Cervical cancer cells were inoculated into 24-well plates. Subsequently, basing on the instructions of EdU kit, EdU solution was loaded into each well and the cells were incubated for 4 h, and then washed by PBS. The cells were fixed with 4% paraformaldehyde. After that, Apollo solution was added to incubate the cells in dark for 30 min, and 0.5% Triton X-100 was added to increase the permeability of the cells. After that, the cells were incubated with Hoechst 33,342. After that, the cells were observed and a fluorescence microscope. Edu positive cell rate = number of red fluorescence-labeled cells/number of blue fluorescence-labeled cells × 100%.
Wound Healing Assay
We used the wound healing assay to evaluate the cell migration ability of cervical cancer cells in the serum-free medium. Hela cells and SiHa cells were transfected with APOC1 siRNA in the six-well plate, after 48 hrs of transfection, we used a white pipette tip to draw a horizontal line in the middle of the six-well plate, then washed the cells with PBS three times to remove floating cells, and then added serum-free medium. Finally, we took pictures with an inverted microscope at 0hrs and 48 hrs after the scratch to evaluate cell wound healing. The relative migration distance of scratches was calculated.
Transwell Assay
We used a 24-well transwell chamber which was equipped with a filter with 8μm pores without Matrigel (Corning Life Sciences, Inc., New York, USA) for the transwell migration experiment. In addition, we used the chamber with Matrigel (Corning Life Sciences, Inc., NY, USA) to evaluate cell invasion ability. We added 500ul of 20% FBS medium to the lower chamber, and added 100ul of serum-free medium containing 5×104 cells to the upper chamber, gently tapped and shaked the plate. The 24-well plate was incubated at 37°C, 5% CO2 for 24 hrs for migration analysis, 48 hrs for invasion analysis, and then the cells were fixed in paraformaldehyde for 20 mins and stained with crystal violet (Beyotime Biotechnology Research Institute, Shanghai, China) 20 mins. Then, we separated the cells on the upper side of the filter with a cotton swab, and washed the filter three times with PBS. Finally, we used an inverted fluorescence microscope (Olympus IX81, Japan) to count the cells at the bottom of the filter.
Western Blot
We used RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) to extract proteins from cultured cervical cancer cells and used a BCA protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China) to determine the concentration of the protein. We then performed SDS-PAGE to separate 15µg of protein lysate. Then incubated PVDF membrane at room temperature with 5% BSA for 1 hr to saturate non-specific sites. Then, the primary antibody, as shown in Table S including APOC1, N-cadherin, Vimentin, Twist, Slug, Snail, CD44, and E-cadherin was incubated overnight at 4°C at the recommended concentration. After incubation with the appropriate HRP-conjugated anti-rabbit or anti-mouse antibody (Amersham Biosciences), the specifically bound protein was detected by ECL according to the manufacturer’s protocol (Amersham Pharmacia Biotech). The density of a specific frequency band is quantified using Image J software. The relative band density is calculated as the ratio of GAPDH.
Co-Immunoprecipitation
After Hela cells reached at least 90% confluence in the 10 cm dishes, they are lysed in 0.5 mL of lysis buffer, and then sonicated and centrifuged at 14,000 g for 10 mins at 4°C. The lysate was incubated with 5µg protein A and 5uL protein G. After incubating with rotation for 1 hr at 4°C, the supernatant was collected. Then, they were incubated with 2ug mouse IgG (Santa Cruze) and 2ug APOC1 antibody (Santa Cruze) with rotation at 4°C overnight. The next day, the beads were collected after protein A/G rotation incubation 3 hrs and centrifuge. Finally, 20uLSDS lysis was added to resolve the beads after washing three times.
Animal Experiments
We used female BALB/c-Nude mice (6–8 weeks old) for in vivo xenograft research. We strictly complied with the approved institutional animal use regulations. For the determination of tumor growth in vivo, Hela-shRNA-Ctrl or Hela-shAPOC1 (2×106) cells were subcutaneously implanted into the left armpits of nude mice or tail vein injection and grown for 4 weeks. We applied a vernier caliper to measure the length and width of the tumor three times every 5 days, and used the formula (length/2) × (width2) to calculate the tumor volume using the average length and width. The experiments have been approved by the Ethics Committee of Soochow University. The animal experiment guide was obeyed to the “Laboratory animal—Guideline for ethical review of animal welfare” (GB/T 35,892–2018).
Statistical Analysis
Every experimental assay was executed in triplicate. Data of those biological replicates or samples were shown as mean±standard deviation (SD). Assays’ statistical analyses were used by SPSS 20.0 software, etc. The relative expression analysis of APOC1 was used by paired samples’ t-test. CCK-8 assay data analysis was used by ANOVA was used to analyze. Other data analysis was used by the independent samples’ t-test. P< 0.05 was regarded as the statistically significant one.
Results
APOC1 Was Up-Regulated in Cervical Cancer
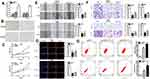
Compared to para-carcinoma specimens, the APOC1 expression was increased in 66.67% (24 of 36) of carcinoma specimens (P <0.001), and about 2.037 times (Figure 1A and B). Scotto Cervix, Zhai Cervix and Biewenga Cervix Statistics database (Figure 1C–E) revealed that the relative expression of APOC1 was up-regulated in cervical carcinomas compared to normal tissues. At the same time, retrospective data analysis showed that the survival cycle analysis was statistically significant (Figure 1F). APOC1 was closely related to Serum SCCA (P<0.001), differentiation (P = 0.002), and FIGO staging (P= 0.016) (Table 1).
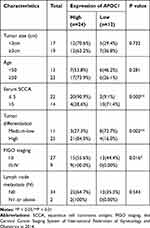 |
Table 1 Correlation Between APOC1 Expression and Clinicopathological Characteristics of Cervical Cancer Patients |
APOC1 Acts as the Oncogene in the Cervical Cancer
The relative expression levels of APOC1 were decreased 76.70% (p=0.0033, si-APOC1-1) and 62.15% (p=0.0068, si-APOC1-2) in HeLa, and 69.93% (p=0.0004, si-APOC1-1) and 66.91% (p<0.001, si-APOC1-2) in SiHa (Figure 2A). The morphology of the Hela and SiHa cells has been changed before and after APOC1 silencing (Figure 2B), which maybe associate the cellular phenotype with the expression of the epithelial and mesenchymal markers. CCK-8 assays showed that knockdown of APOC1 (Figure 2C) significantly restrained cell proliferation in cervical cancer cells (p < 0.01 in cervical cancer cell lines). Compared to control group, EdU assays elucidated that the EdU positive cells quantity in si-APOC1 group was decreased by 64.27% in HeLa (P = 0.0028, si-APOC1-1) and by 54.72% in HeLa (P = 0.0022, si-APOC1-2) (Figure 2D), EdU assays elucidated that the EdU positive cells quantity in si-APOC1 group was decreased by 75.26% in SiHa (P = 0.0066, si-APOC1-1) and by 67.53% in SiHa (P = 0.0107, si-APOC1-2) (Figure 2D). These data illustrated that knockdown of APOC1 restrained cell proliferation in cervical cancer cell lines.
Cells scratch assays were used to detect the plasmids' role in cell migration. Scratch assays elucidated that the ratio of the relative migration in si-APOC1 group was decreased by 69.96% in HeLa (P = 0.0062, si-APOC1-1) and by 56.79% in HeLa (P = 0.0132, si-APOC1-2) (Figure 2E). Scratch assays elucidated that the ratio of the relative migration in si-APOC1 group was decreased by 68.52% in SiHa (P = 0.0063, si-APOC1-1) and by 62.90% in SiHa (P = 0.0165, si-APOC1-2) (Figure 2E). Transwell assays elucidated that the ratio of the relative migration was decreased by 73.16% in HeLa (P = 0.0090, si-APOC1-1) and 49.11% in HeLa (P = 0.0242, si-APOC1-2) and decreased by 74.25% in SiHa (P =0.0056, si-APOC1-1) and 62.88% in SiHa (P = 0.0189, si-APOC1-2) (Figure 2F), and the ratio of the relative invasion was decreased by in si-APOC1 group was decreased by 67.60% in HeLa (P = 0.0036, si-APOC1-1) and by 55.94% in HeLa (P = 0.0058, si-APOC1-2), and decreased by 64.82% in SiHa (P = 0.0009, si-APOC1-1) and 49.60% in SiHa (P = 0.0021, si-APOC1-2) (Figure 2F). These results illustrated that knockdown of APOC1 restrained cervical cancer cells migration and invasion. Flow cytometry assays were used to detect cell apoptosis. The ratios of apoptosis in cervical cancer cells were up-regulated after transfection with si-APOC1. Compared to NC, the ratios of apoptosis were up-regulated by 10.69 times in HeLa (P< 0.001, si-APOC1-1) and 7.482 times in Hela (P< 0.001, si-APOC1-2) (Figure 2G) after transfection siAPOC1, the ratios of apoptosis were up-regulated by 8.338 times in SiHa (P = 0.0185, si-APOC1-1) and 4.088 times in SiHa (P < 0.001, si-APOC1-2) (Figure 2G) after transfection siAPOC1. In brief, knockdown of APOC1 promoted cervical cancer cell apoptosis.
Over-Expression of APOC1 Promoted Cell Progression of Cervical Cancer Cell Lines
The relative APOC1 expression levels were increased about 8.761 times (P = 0.0002) in HeLa and 5.171 times (P= 0.0011) in SiHa (Figure 3A). CCK-8 assays showed that over-expression of APOC1 (Figure 3B) significantly promoted cell proliferation in cervical cancer cells (p < 0.01).
Compared to the control group, EdU assays elucidated that the EdU positive cells’ quantity in pcDNA3.1-APOC1 group was obviously up-regulated about 4.190 times in HeLa (P < 0.0001) (Figure 3C), and about 7.286 times in SiHa (P=0.0037) (Figure 3C). The data illustrated that over-regulation of APOC1 accelerated cell proliferation in cervical cancer cell lines.
Scratch assays elucidated that the ratio of the relative migration in pcDNA3.1-APOC1 group was up-regulated about 4.472 times in HeLa (P = 0.0002) (Figure 3D), and up-regulated about 3.103 times in SiHa (P = 0.0038) (Figure 3D). These results illustrated that over-regulation of APOC1 promoted cervical cancer cells’ migration.
Transwell assays elucidated that the ratio of the relative migration in pcDNA3.1-APOC1 group was increased about 2.328 times in HeLa (P = 0.0080) (Figure 3E) and 2.794 times in SiHa (P = 0.0105) (Figure 3E). These results illustrated that over-regulation of APOC1 promoted cervical cancer cells. Transwell assays elucidated that the ratio of the relative invasion in pcDNA3.1-APOC1 group was up-regulated about 2.349 times in HeLa (P = 0.0002) (Figure 3E) and 2.197 times in SiHa (P = 0.0168) (Figure 3E). These results illustrated that over-regulation of APOC1 promoted cervical cancer cells’ migration.
The ratios of apoptosis in cervical cancer cells were down-regulated after transfection with pcDNA3.1-APOC1. Compared to control groups, the ratios of apoptosis were down-regulated by 83.05% in HeLa (P= 0.0015) and 68.69% times in Hela (P= 0.0004) (Figure 3F) after transfection pcDNA3.1-APOC1.
APOC1 is Involved in EMT Signal Axis
IP pyrolysis products are used for proteomics detection (Figure 4A–F), which was furthermore proved that APOC1 bound multiple functional proteins in cervical cancer cells, and KEGG, GO and other analyses were performed, and that further proved that APOC1 could interact with many proteins and may play an important function in cervical cancer cells. CD44 was confirmed to be an APOC1-binding protein in the Hela cell line by immunoprecipitation (Figure 4G). Down-regulated APOC1 could reduce APOC1 expression and restrained APOC1 could refer to EMT signaling in cervical cancer cells (Figure 4H). Western blotting was used to detect the EMT signaling-associated downstream genes expression. Knockdown of APOC1 decreased N-cadherin, Vimentin, Twist, Slug, Snail and CD44 expression and increased E-cadherin expression in cervical cancer cells.
Knockdown of APOC1 Restrained Tumorigenicity of Cervical Cancer Cells
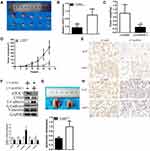
Generation of xenograft showed that knockdown of APOC1 restrained the tumorigenicity of cervical cancer cells in vivo (Figure 5A–E). Tumors collected from mice were shown in Figure 5A. It’s shown that knockdown of APOC1 was down-regulated compared to LV-shNC of cervical cancer cells in vivo (Figure 5B). Compared to the LV-negative control treatment group, tumor weight was less in the LV-shAPOC1 group (Figure 5C). Tumor growth of LV-shNC was faster than the LV-shAPOC1 group (Figure 5D). Meanwhile, knockdown of APOC1 down-regulated Ki-67 and APOC1 expression (Figure 5E) of cervical cancer cells in vivo. These data manifested that APOC1 promoted tumorigenicity of cervical cancer cells in vivo. Knockdown of APOC1 decreased N-cadherin, Vimentin and CD44 expression and increased E-cadherin expression of cervical cancer cells in vivo (Figure 5F). Knockdown of APOC1 significantly reduced the number and size of pulmonary metastases (Figure 5G). Moreover, differential expression of Ki-67 and APOC1 in metastatic specimens (Figure 5H). The results indicated that APOC1 promoted cervical carcinoma metastasis in vivo.
Discussion
Cervical cancer, as one of the common human tumors, is one of the most common gynecological malignancies,1–5,34 which is like other tumors35–41 and they threatened seriously to human health as well as cervical cancer,42–48 its morbidity and mortality rate are at the forefront of female malignancies. Surgery, radiotherapy and chemotherapy for early cervical cancer have good effects, but in advanced stage, the therapeutic effect of metastatic and recurrent cervical cancer is very limited and the survival rate is low.6–10 How to effectively treat advanced and recurrent cervical cancers is the hot spot of current research. In this study, we proved the correlation between APOC1 and cervical cancer through a large number of experiments and data analysis, and provided a new target for cervical cancer. Of course, more specific mechanisms needed to be verified through further research.
The basic functions of apolipoprotein include the delivery of lipids, the activation of lipoprotein metabolic enzymes, and the recognition of receptors. APOC1 genes are expressed mostly in the liver, more frequently in the lungs, skin, testicles, and spleen and so on. APOC1, also known as Apo-CI; ApoC-I; apo-CIB; apoC-IB, is a new biomarker amplified by 19q13.32 and this gene encoded a member of the apolipoprotein C1 family.11,49–51 The gene is expressed primarily in the liver, and it is activated when monocytes differentiate into macrophages. The encoded protein plays a central role in high-density lipoprotein (HDL) and very low-density lipoprotein (VLDL) metabolism.47 According to the results of previous studies, APOC1 was involved in the progression and development of various cancers. APOC1 was abnormally expressed in gastric cancer (GC), colorectal cancer (CRC), prostate cancer (PCa), lung cancer, pancreatic cancer, renal cancer and so on.26–31
APOC1 was significantly higher in GC and associated with clinical stage, tumor classification and the lymph node metastasis and overall survival.26 APOC1 was up-regulated in CRC tissues and tight related to poor prognosis. APOC1 expression regulated CRC progression via MAPK axis.27 Knockdown of APOC1 restrained cell progression in PCa.28 There was a positive correlation with the expression levels of IL-6 and APOC1 in lung cancer progression.29 The expression of APOC1 tightly related to the poor prognosis in pancreatic preoperative serum and knockdown of APOC1 restrained cell progression.30 ApoC1 related to poor survival and promoted progression via EMT pathway and activation of STAT3 in renal cancers.31 In a word, APOC1 plays an important role in many cancers. In this experiment, we intended to study APOC1 function and underlying mechanism in cervical cancers, we found that APOC1 accelerated the progression of cervical cancer via EMT. We found that the relative expression of APOC1 was up-regulated in cervical cancer tissues. Knockdown of APOC1 restrained cervical cancer cells’ progression and over-expression of APOC1 accelerated cervical cancer cells’ progression. Furthermore, studies showed that APOC1 impeding inhibited tumor cell growth in vivo. We inferred that APOC1 played an important role in the progression and metastasis of cervical cancers and may participate in its progression mechanism, and provided us with direction for clinical therapy, and even provided a direction for the effective treatment of advanced and recurrent cervical cancer. On the basis of the results above, APOC1 could be an important therapeutic target and biological marker for cervical cancer in future.
Funding
This work was supported by National Natural Science Foundation of China (No.81672560, 81772773 and 81302275), Jiangsu Provincial Medical Youth Talent (No. QNRC2016753), Suzhou Clinical Key Technology Project (No. LCZX201705), Gusu Medical Youth Talent (No. GSWS2019034), The Project of Jiangsu Provincial Maternal and Child Health Association (No. FYX201709) and Suzhou Clinical Trial Project (No. SLT201912).
Disclosure
All authors declared that they have no conflicts of interests.
References
1. Casullo A, Buonaguro L, Buonaguro FM, Tornesello ML. The role of RNA splicing factors in cancer: regulation of viral and human gene expression in human papillomavirus-related cervical cancer. Front Cell Dev Biol. 2020;8:474. doi:10.3389/fcell.2020.00474.
2. Zhang C, Hua Y, Qiu H, et al. KMT2A regulates cervical cancer cell growth through targeting VDAC1. Aging. 2020;12(10):9604–9620. doi:10.18632/aging.103229
3. Wu H, Song S, Yan A, et al. RACK1 promotes the invasive activities and lymph node metastasis of cervical cancer via galectin-1. Cancer Lett. 2020;469:287–300. doi:10.1016/j.canlet.2019.11.002.
4. Tran AM, Chalbatani GM, Berland L, et al. A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Front Cell Dev Biol. 2020;8(50). doi:10.3389/fcell.2020.00050.
5. Wang AH, Jin CH, Cui GY, et al. MIR210HG promotes cell proliferation and invasion by regulating miR-503-5p/TRAF4 axis in cervical cancer. Aging. 2020;12(4):3205–3217. doi:10.18632/aging.102799.
6. Chen P, Zhao X, Zhou F, et al. Characterization of 500 Chinese patients with cervical esophageal cancer by clinicopathological and treatment outcomes. Cancer Biol Med. 2020;17(1):219–226. doi:10.20892/j.issn.2095-3941.2019.0268.
7. Wang R, Pan W, Jin L, et al. Human papillomavirus vaccine against cervical cancer: opportunity and challenge. Cancer Letters. 2020;471:88–102. doi:10.1016/j.canlet.2019.11.039
8. Chen R, Chen Y, Zhao W, et al. The role of methyltransferase nsd2 as a potential oncogene in human solid tumors. Onco Targets Ther. 2020;13:6837–6846. doi:10.2147/OTT.S259873.
9. Li N, Cheng C, Wang T. MiR-181c-5p mitigates tumorigenesis in cervical squamous cell carcinoma via targeting glycogen synthase kinase 3β interaction protein (GSKIP). Onco Targets Ther. 2020;13:4495–4505. doi:10.2147/OTT.S245254.
10. Zhao J, Yang T. LncRNA FOXP4-As1 is involved in cervical cancer progression via regulating mir-136-5p/cbx4 axis. Onco Targets Ther. 2020;13:2347–2355. doi:10.2147/OTT.S241818.
11. Fuior EV, Gafencu AV. Apolipoprotein c1: its pleiotropic effects in lipid metabolism and beyond. Int J Mol Sci. 2019;20(23):5939. doi:10.3390/ijms20235939.
12. Zhou X, Chen Y, Mok KY, et al. Non-coding variability at the APOE locus contributes to the Alzheimer’s risk. Nat Commun. 2019;10(1):3310. doi:10.1038/s41467-019-10945-z.
13. Guo Y, Xu W, Li JQ, et al. Genome-wide association study of hippocampal atrophy rate in non-demented elders. Aging. 2019;11(22):10468–10484. doi:10.18632/aging.102470.
14. Xing QQ, Liu LW, Zhao X, Lu Y, Dong YM, Liang ZQ. Serum proteomics analysis based on label-free revealed the protective effect of Chinese herbal formula Gu-Ben-Fang-Xiao. Biomed Pharmacother. 2019;119:109390. doi:10.1016/j.biopha.2019.109390.
15. Wang W, Wang B, Liu C, et al. Serum proteomic predicts effectiveness and reveals potential biomarkers for complications in liver transplant patients. Aging. 2020;12(12):12119–12141. doi:10.18632/aging.103381.
16. Li HZ, Xu XH, Lin N, et al. Overexpression of miR-10a-5p facilitates the progression of osteoarthritis. Aging. 2020;12(7):5948–5976. doi:10.18632/aging.102989.
17. Han C, He Y, Chen L, et al. Low expression of APOB mRNA or its hypermethylation predicts favorable overall survival in patients with low-grade glioma. Onco Targets Ther. 2020;13:7243–7255. doi:10.2147/OTT.S257794.
18. Huang CC, Charng MJ. Genetic diagnosis of familial hypercholesterolemia in Asia. Front Genet. 2020;11:833. doi:10.3389/fgene.2020.00833.
19. Sun Y, Hou XH, Wang DD, et al. Apolipoprotein B/AI ratio as an independent risk factor for intracranial atherosclerotic stenosis. Aging. 2019;11(17):6851–6862. doi:10.18632/aging.102216.
20. Kuo CL, Pilling LC, Atkins JL, Kuchel GA, Melzer D. ApoE e2 and aging-related outcomes in 379,000 UK Biobank participants. Aging. 2020;12(12):12222–12233. doi:10.18632/aging.103405.
21. Yang Y, Tang F, Wei F, et al. Silencing of long non-coding RNA H19 downregulates CTCF to protect against atherosclerosis by upregulating PKD1 expression in ApoE knockout mice. Aging. 2019;11(22):10016–10030. doi:10.18632/aging.102388.
22. Li Q, Wang Q, Xu W, et al. C-reactive protein causes adult-onset obesity through chronic inflammatory mechanism. Front Cell Dev Biol. 2020;8:18. doi:10.3389/fcell.2020.00018
23. Guo M, Xiao J, Sheng X, et al. Ginsenoside rg3 mitigates atherosclerosis progression in diabetic ApoE-/- mice by skewing macrophages to the m2 phenotype. Front Pharmacol. 2018;9:464. doi:10.3389/fphar.2018.00464
24. Famenini S, Rigali EA, Olivera-Perez HM, et al. Increased intermediate M1-M2 macrophage polarization and improved cognition in mild cognitive impairment patients on ω-3 supplementation. FASEB J. 2017;31(1):148–160. doi:10.1096/fj.201600677RR
25. Shi X, Ma W, Li Y, et al. MiR-144-5p limits experimental abdominal aortic aneurysm formation by mitigating M1 macrophage-associated inflammation: suppression of TLR2 and OLR1. J Mol Cell Cardiol. 2020;143:1–14. doi:10.1016/j.yjmcc.2020.04.008
26. Yi J, Ren L, Wu J, et al. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for gastric cancer. Ann Transl Med. 2019;7(16):380. doi:10.21037/atm.2019.07.59
27. Ren H, Chen Z, Yang L, et al. Apolipoprotein C1 (APOC1) promotes tumor progression via MAPK signaling pathways in colorectal cancer. Cancer Manag Res. 2019;11:4917–4930. doi:10.2147/CMAR.S192529
28. Su WP, Sun LN, Yang SL, et al. Apolipoprotein C1 promotes prostate cancer cell proliferation in vitro. J Biochem Mol Toxicol. 2018;32(7):e22158. doi:10.1002/jbt.22158
29. Ko HL, Wang YS, Fong WL, Chi MS, Chi KH, Kao SJ. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker Phase I trial. Thorac Cancer. 2014;5(6):500–508. doi:10.1111/1759-7714.12117
30. Takano S, Yoshitomi H, Togawa A, et al. Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene. 2008;27(20):2810–2822. doi:10.1038/sj.onc.1210951
31. Li YL, Wu LW, Zeng LH, et al. ApoC1 promotes the metastasis of clear cell renal cell carcinoma via activation of STAT3. Oncogene. 2020. doi:10.1038/s41388-020-01428-3
32. Chen M, Li J, Zhuang C, Cai Z. Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of bladder cancer. Oncotarget. 2017;8(17):28176–28186. doi:10.18632/oncotarget.14945
33. Chen M, Zhuang C, Liu Y, et al. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 2016;376(1):155–164. doi:10.1016/j.canlet.2016.03.037
34. Sun Z, Niu S, Xu F, et al. CircAMOTL1 promotes tumorigenesis through miR-526b/SIK2 axis in cervical cancer. Front Cell Dev Biol. 2020.8;569190. doi:10.3389/fcell.2020.568190.
35. Chen M, Wei X, Shi X, et al. LncRNA HIF1A-AS2 accelerates malignant phenotypes of renal carcinoma by modulating miR-30a-5p/SOX4 axis as a ceRNA. Cancer Biol Med. 2020. doi:10.20892/j.issn.2095-3941.2020.0209.
36. Hu X, Yang D, Li Y, et al. Erratum to Prevalence and clinical significance of pathogenic germline BRCA1/2 mutations in Chinese non-small cell lung cancer patients. Cancer Biol Med. 2020;17(2):513. doi:10.20892/j.issn.2095-3941.2020.0029
37. Yao C, Su L, Zhang F, et al. Thevebioside, the active ingredient of traditional Chinese medicine, promotes ubiquitin-mediated SRC-3 degradation to induce NSCLC cells apoptosis. Cancer Lett. 2020:
38. Gao T, Yu L, Fang Z, et al. KIF18B promotes tumor progression in osteosarcoma by activating β-catenin. Cancer Biol Med. 2020;17(2):371–386. doi:10.20892/j.issn.2095-3941.2019.0452
39. Meng LD, Shi GD, Ge WL, et al. Linc01232 promotes the metastasis of pancreatic cancer by suppressing the ubiquitin-mediated degradation of HNRNPA2B1 and activating the A-Raf-induced MAPK/ERK signaling pathway. Cancer Lett. 2020:
40. Li Y, Xin J, Sun Y, Han T, Zhang H, An F. Magnetic resonance imaging-guided and targeted theranostics of colorectal cancer. Cancer Biol Med. 2020;17(2):307–327. doi:10.20892/j.issn.2095-3941.2020.0072
41. Huang X, Reye G, Momot KI, et al. Heparanase promotes syndecan-1 expression to mediate fibrillar collagen and mammographic density in human breast tissue cultured ex vivo. Front Cell Dev Biol. 2020;8:599. doi:10.3389/fcell.2020.00599
42. Wang Z,Jiang Q, Dong C. Metabolic reprogramming in triple-negative breast cancer. Cancer Biol Med. 2020;17(1):44–59. doi:10.20892/j.issn.2095-3941.2019.0210
43. Melissari MT, Chalkidi N, Sarris ME, Koliaraki V. Fibroblast reprogramming in gastrointestinal cancer. Front Cell Dev Biol. 2020;8:630. doi:10.3389/fcell.2020.00630
44. Lan Y, Lou J, Hu J, Yu Z, Lyu W, Zhang B. Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway. Cancer Biol Med. 2020;17(1):112–131. doi:10.20892/j.issn.2095-3941.2019.0164
45. Teo WS, Holliday H, Karthikeyan N, et al. Id proteins promote a cancer stem cell phenotype in mouse models of triple negative breast cancer via negative regulation of robo1. Front Cell Dev Biol. 2020;8:552. doi:10.3389/fcell.2020.00552
46. Zhao C, Meng X, Li L, et al. Neutrophil extracellular traps mediate the crosstalk between glioma progression and the tumor microenvironment via the HMGB1/RAGE/IL-8 axis. Cancer Biol Med. 2020;17(1):154–168. doi:10.20892/j.issn.2095-3941.2019.0353
47. Zeng J, Li G, Xia Y, et al. miR-204/COX5A axis contributes to invasion and chemotherapy resistance in estrogen receptor-positive breastCancers.Cancer Lett. 2020:
48. Sun X, Wang M, Wang M, et al. Exploring the metabolic vulnerabilities of epithelial-mesenchymal transition in breast cancer. Front Cell Dev Biol. 2020;8:655. doi:10.3389/fcell.2020.00655
49. Zhou Q, Peng D, Yuan X, et al. APOE and APOC1 gene polymorphisms are associated with cognitive impairment progression in Chinese patients with late-onset Alzheimer’s disease. Neural Regen Res. 2014;9(6):653–660. doi:10.4103/1673-5374.130117
50. Zhang R, Liu Q, Liu H, et al. Effects of apoC1 genotypes on the hormonal levels, metabolic profile and PAF-AH activity in Chinese women with polycystic ovary syndrome. Lipids Health Dis. 2018;17(1):77. doi:10.1186/s12944-018-0725-5
51. Li J,Sun L, Xu F, et al. Screening and Identification of APOC1 as a novel potential biomarker for differentiate of mycoplasma pneumoniae in children. Front Microbiol. 2016;7:1961. doi:10.3389/fmicb.2016.01961
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.