Back to Journals » International Journal of Nanomedicine » Volume 17
Antisense vicR-Loaded Dendritic Mesoporous Silica Nanoparticles Regulate the Biofilm Organization and Cariogenicity of Streptococcus mutans
Authors Tian Y , Zhang Y, Zhang M, Chen X, Lei L , Hu T
Received 18 August 2021
Accepted for publication 9 February 2022
Published 21 March 2022 Volume 2022:17 Pages 1255—1272
DOI https://doi.org/10.2147/IJN.S334785
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ebrahim Mostafavi
Yuting Tian,1,* Yue Zhang,1,* Mengjiao Zhang,2 Xianchun Chen,3 Lei Lei,1 Tao Hu1
1State Key Laboratory of Oral Diseases, Department of Preventive Dentistry, West China Hospital of Stomatology, Sichuan University, Chengdu, Sichuan, 610041, People’s Republic of China; 2West China School of Pharmacy, Sichuan University, Chengdu, 610041, People’s Republic of China; 3College of Polymer Science and Engineering, Sichuan University, Chengdu, 610065, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xianchun Chen; Tao Hu, Tel +86 13880649396, +86 28-85503486, Email [email protected]; [email protected]
Purpose: VicR is the essential response regulator related to the synthesis of exopolysaccharide (EPS) – one of the main cariogenic factors of S. mutans. An antisense vicR RNA (ASvicR) could bind to vicR mRNA, hindering the transcription and translation of the vicR gene. We had constructed a recombinant plasmid containing the ASvicR sequence (plasmid-ASvicR) and proved that it could reduce EPS synthesis, biofilm formation, and cariogenicity. However, the recombinant plasmids are supposed to be protected from enzymatic degradation and possess higher transformation efficiency. The principal objective of the present research was to construct an appropriate vector that can carry and protect the plasmid-ASvicR and investigate the effects of the carried plasmids on the cariogenicity of the S. mutans.
Methods: Aminated dendritic mesoporous silica nanoparticles (DMSNs-NH2) were synthesized and characterized. The ability of DMSNs-NH2 to carry and preserve the plasmid-ASvicR (DMSNs-NH2-ASvicR) was proved by the loading curve, agarose electrophoresis, DNase I digestion assays, and energy-dispersive spectrometry (EDS) mapping. Transformation assays demonstrated whether the plasmid could enter S. mutans. The effect of DMSNs-NH2-ASvicR on the 12-hour and 24-hour biofilms of S. mutans was evaluated by biofilm formation experiments and quantitative reverse transcription polymerase chain reaction (qRT-PCR). The cytotoxicity of DMSNs-NH2-ASvicR was assessed by CCK-8 and live/dead staining assays. The regulation of DMSNs-NH2-ASvicR on the cariogenicity of S. mutans was also evaluated in vivo.
Results: DMSNs-NH2 could load approximately 92% of plasmid-ASvicR at a mass ratio of 80 and protect most of plasmid-ASvicR from degradation by DNase I. The plasmid-ASvicR loaded on DMSNs-NH2 could be transformed into S. mutans, which down-regulated the expression of the vicR gene, reducing EPS synthesis and biofilm organization of S. mutans. DMSNs-NH2-ASvicR exhibited favorable biocompatibility, laying a foundation for its subsequent biomedical application. In addition, DMSNs-NH2-ASvicR led to decreased caries in vivo.
Conclusion: DMSNs-NH2 is a suitable vector of plasmid-ASvicR, and DMSNs-NH2-ASvicR can inhibit biofilm formation, reducing the cariogenicity of S. mutans. These findings reveal that DMSNs-NH2-ASvicR is a promising agent for preventing and treating dental caries.
Keywords: biofilm, exopolysaccharides, cariogenicity, mesoporous silica nanoparticles, Streptococcus mutans
Introduction
Dental caries accounts for the highest incidence of oral disease, causing a significant disease burden in many countries.1 The leading etiological agent of dental caries is Streptococcus mutans (S. mutans), which can form biofilms on solid surfaces such as the enamel, tooth root surfaces, or dental implants.2,3 Biofilms are three-dimensional structures consisting of microbes and an extracellular matrix. The exopolysaccharides (EPS) produced by the interactions of glucosyltransferases (Gtfs) and fructosyltransferases (Ftfs) with sucrose in S. mutans are the main components and the essential virulence factors of the dental caries.4–6 The EPS matrix enhances the adhesion of microorganisms, promotes the biofilm formation of three-dimensional structures to impede the penetration of antibiotic substances into the biofilm, and plays a role as a source of energy.4,5,7 Therefore, reducing the synthesis of the EPS matrix could decrease the virulence of S. mutans, lowering the incidence of caries.
Two-component signal transduction systems (TCSTS) could modulate gene expression in response to external environmental changes.8 There are thirty TCSTS in S. mutans based on their genomic sequence.9 The VicRK system, one of the TCSTS, consists of a membrane-bound sensor (VicK) and a cytoplasmic response regulator (VicR). It has been reported that VicR regulated the expression of genes that are associated with EPS synthesis.8,10,11 However, the inability to construct vicR deletion mutants revealed that the vicR gene was also crucial to the viability of S. mutans.8 Previous reports indicated that antisense (AS) substances could inhibit the transcription and translation of the related genes.12–14 An AS RNA could bind to complementary mRNA via Watson-Crick base pairing and suppress its transcription by inducing a steric blockage and/or RNase degradation.15–17 Our previous studies have shown that an antisense vicR RNA (ASvicR) could be complementary to the vicR mRNA through base pairing, forming an ASvicR-vicR duplex structure and hindering the expression of the vicR gene.18 In addition, ASvicR inhibited EPS synthesis, bacterial growth, and cariogenicity of S. mutans, indicating that ASvicR could be considered a potential strategy for caries prevention.18,19 We have constructed a recombinant plasmid containing the ASvicR gene (plasmid-ASvicR) by linking the ASvicR sequence to the pDL278 plasmid.18 However, the plasmids are supposed to be protected from enzymatic degradation and avoid electrostatic repulsion from the cell surface to endow the recombinant plasmid with higher transformation efficiency.17,20,21 Hence, it is necessary to explore an appropriate vector that can protect recombinant plasmids and promote their transformation.
Various strategies have been demonstrated to be nucleic acid carriers, including viruses,22 lipids,23 peptides,24 cationic polymers,25 and inorganic nanoparticles.26,27 Despite their high transformation efficiency, the biocompatibility and safety of viral vectors remain controversial.21,26,28 In addition, the immunogenicity of liposomes, the poor delivery efficiency of peptides, and the cytotoxicity of cationic polymers still require further studies to determine whether these materials are suitable as gene delivery vectors.20,29,30 Among the inorganic nanoparticles, mesoporous silica nanoparticles (MSNs) exhibit excellent potentials as drug and gene delivery vectors due to their favorable biocompatibility, chemical stability, large surface area, and simple synthesis process.31–34 Furthermore, silica has been classified as generally recognized as safe (GRAS) by the US Food and Drug Administration (FDA) and is commonly used as a food additive as well as in the cosmetic and pharmaceutical industries.35 However, the pore sizes of several conventional MSNs are too small to accommodate and protect macromolecule such as nucleic acids and proteins.36–38 Improvements in the synthesis methods have resulted in the emergence of different morphologies (spherical, sheet, worm-like, rod-like, fibrous, dendritic, etc) and architectures (solid, hollow, core-shell, Janus, yolk-shell, etc.) in MSNs.39,40 In recent years, dendritic mesoporous silica nanoparticles (DMSNs) with center-radial oriented pore structures have aroused great interest due to their higher pore permeability, larger pore volume, and better accessibility to the particle inner surface.41–44 There are several reports about MSNs acting as gene carriers in different cells. However, for bacteria, MSNs are mainly utilized to load traditional antibiotics, metallic ions, proteins, and plant extracts,45–49 which react on a wide range of bacteria, leading to drug resistance and dysbacteriosis. To load the plasmid-ASvicR and act on the bacteria, the DMSNs should have a positively charged surface, a larger pore size, and a smaller particle size.
Based on the facts mentioned above, we hypothesized that dendritic mesoporous silica nanoparticles could be vectors of plasmid-ASvicR to target the transcriptions and translations of vicR genes to reduce the EPS synthesis in the biofilm and cariogenicity of S. mutans. The delivery system is supposed to have the excellent ability to carry and protect plasmids. The novel plasmid-loaded DMSNs that could target S. mutans show potential as a method of dental caries management.
Materials and Methods
Materials
Cetyltrimethylammonium chloride (CTAC), triethanolamine (TEA), tetraethyl orthosilicate (TEOS), cyclohexane, and 3-aminopropyltriethoxysilane (APTES) were purchased from Macklin (Shanghai, China). Hydrochloric acid (37%) and anhydrous ethanol were purchased from Kelong (Chengdu, China). EndoFree Maxi Plasmid Kit was purchased from Tiangen (Beijing, China). Brain Heart Infusion (BHI) medium was purchased from Oxiod (USA). Alexa Fluor® 647 and SYTO™ 9 dyes were purchased from Life Tech (USA). MasterPure™ RNA purification kit was purchased from Epicentre (USA). DNase I, PrimeScript™ RT reagent kit with gDNA Eraser, and TB Green® Premix Ex Taq™ II were purchased from Takara (Japan). Cell counting kit-8 (CCK-8) was purchased from Dojindo (Japan). Live/dead cell double staining kit (Calcein-AM/PI) was purchased from Solarbio (Beijing, China). DMEM, FBS, trypsin, and PBS were purchased from ThermoFisher (USA). Keyes2000# cariogenic fodder was purchased from Dossy (Chengdu, China).
Synthesis of DMSNs
DMSNs were synthesized according to previous literature with some modifications.42,50,51 Six gram of CTAC and 0.18 g of TEA were added to 60 mL of ultrapure water. Then, the solutions were stirred (300 rpm) in a 100-mL round-bottom flask at 60°C for 1 hour. Next, 20 mL of TEOS in cyclohexane (20 v/v%) was carefully added to the water-CTAC-TEA solution and kept at 60°C in an oil magnetic stirrer for 10 hours. The products were collected by centrifugation, washed with ethanol three times, added to 100 mL of 1 v/v% hydrochloric acid ethanol solution, and refluxed at 80°C for 12 hours twice. The final products were washed with ethanol and ultrapure water three times and lyophilized for further investigations.
Surface Amination of DMSNs with APTES
DMSNs were modified to acquire a positively charged surface to load negatively charged plasmids.52,53 In the present study, the amino groups were introduced using 3-aminopropyltriethoxysilane (APTES) through a post-grafting method to obtain positively charged DMSNs.37,54 To prepare amino-DMSNs (DMSNs-NH2), 500 mg of DMSNs was dispersed in 100 mL of ethanol by ultrasonication. Then, 1.5mL of APTES was added to the suspension and the mixtures were refluxed at 80°C for 24 h. The final products were washed with ethanol and ultrapure water three times and lyophilized for the following experiments.
Characterization
Transmission electron microscopy (TEM) (Tecnai G2 F20 S-TWIN; FEI, USA) was used to observe the structure of DMSNs-NH2, and ImageJ software (National Institute of Mental Health, USA) was used to obtain the diameter distribution of the particles. Nitrogen adsorption-desorption measurements were conducted with a specific surface area analyzer (Kubo-X1000; Builder, China) to evaluate the surface areas, pore size distributions, and pore volumes. Zeta potentials were analyzed with a Zetasizer Nano ZS instrument (Malvern, UK), and the measurements were repeated three times. Energy-dispersive spectrometry (EDS) mapping (JEM-F200_TFEG, Japan), and Fourier transform infrared (FTIR) spectroscopy (Thermo Fisher Nicolet Is5, USA) were also conducted to characterize the DMSNs-NH2.
Binding of Plasmid-ASvicR to DMSN-NH2 (DMSNs-NH2-ASvicR)
The recombinant plasmids containing the ASvicR consequence were obtained from E. coli using an EndoFree Maxi Plasmid Kit, and the concentrations of plasmids were measured with NanoDrop Onec (Thermo Fisher, USA). DMSNs-NH2 was suspended in deionized water and prepared into several solutions of 0, 1, 2, 3, 4, 5, 6, 7, 8, 9 μg/μL, respectively. Then, 1 μg of plasmid-ASvicR was dispersed in 10 μL of DMSNs-NH2 solutions of the different concentrations, in which the DMSNs-NH2 was mixed with plasmid-ASvicR by mass at 0, 10, 20, 30, 40, 50, 60, 70, 80, and 90 ratios. After shaking (200 rpm) at room temperature for 1 h, the mixtures were used to perform gel electrophoresis by loading onto 1% agarose gel at 100V for 30 minutes. The resulting products were observed using a Bio-Rad electrophoresis system (Bio-Rad Laboratories, USA).
To quantify the amount of plasmid-ASvicR absorbed on DMSNs-NH2, 6 μg of plasmid-ASvicR was dispersed in 60 μL of DMSNs-NH2 solutions with different concentrations (as mentioned above). After shaking (200rpm) at room temperature for 1 h, the mixtures were centrifuged at 12,000 rpm for 10 min, and the concentration of plasmid-ASvicR in the supernatant was analyzed by NanoDrop Onec. The amount of plasmid-ASvicR adsorbed on DMSNs-NH2 was calculated by subtracting the content in the supernatants from the initial concentration.
In addition, the DMSNs-NH2-ASvicR solution (the weight ratio of DMSNs-NH2/plasmid-ASvicR=80) was prepared as mentioned above. Then the solution was centrifuged at 12,000 rpm for 10 min to obtain DMSNs-NH2-ASvicR precipitate. The precipitate was lyophilized and stored at room temperature for the following experiments. To further prove that plasmids-ASvicR had been absorbed by the DMSNs-NH2, the lyophilized powder of DMSNs-NH2-ASvicR was dispersed in deionized water and used for Zeta potential test, EDS-mapping, and FTIR spectroscopy.
DNase I Digestion Assay
The DMSNs-NH2 mixed with plasmid-ASvicR was prepared by mass ratios of 0, 10, 20, 30, 40, 50, 60, 70, 80, and 90 and incubated with 1 μL of DNase I. After incubation, half of the mixtures were used for 1% agarose gel electrophoresis, as mentioned above. The other half of the mixtures were added to 500-μM EDTA to stop the digestion, and then heparin (4 mg/mL) was added to release the plasmids.37,55 After shaking at 50°C and 200 rpm for 1 h, the mixture was centrifuged, and the supernatant was used for 1% agarose gel electrophoresis.
Transformation of DMSNs-NH2-ASvicR
The plasmid-ASvicR contains the antisense vicR sequence, the spectinomycin-resistant sequence, and the gene encoding the green fluorescent protein (GFP). DMSNs-NH2 was mixed with plasmid-ASvicR at a mass ratio of 80, and the mixture was stored in a refrigerator at 4°C for subsequent experiments. We conducted antibiotic plate screening assay and GFP expression assay to prove that the plasmid-ASvicR loaded on DMSNs-NH2 can enter S. mutans and be expressed.
A single colony of S. mutans UA159 was selected and cultured overnight in a fresh Brain Heart Infusion (BHI) medium at 37°C (80% N2, 10% CO2, and 10% H2). On the one hand, the bacterial suspensions were diluted in 10 mL of fresh BHI medium at a volume ratio of 1:20 and cultured for 2–3 h. After adjusting the optical density at 600nm (OD600) to 0.4, 500 μL of bacterial suspensions were cultured in the presence of deionized water, plasmid-ASvicR, and DMSNs-NH2-ASvicR (containing 200 ng of plasmids-ASvicR) for 30 min, 3 h, 6 h, 12 h, and 24 h, respectively. Then, the mixed solutions were spread onto the BHI solid medium with spectinomycin (Spe, 1mg/mL). On the other hand, the bacterial suspensions cultured overnight were diluted in 10 mL of BHI medium containing 1% sucrose (BHIS) at a volume ratio of 1:10. Then, 2 mL of the diluted liquid was added to the confocal dish, followed by adding deionized water, pDL278 (the plasmid without GFP and antisense vicR sequence), DMSNs-NH2, plasmids-ASvicR, and DMSNs-NHs-ASvicR, respectively. The mixed solutions were cultured for 3 h, and the cultures were evaluated under a confocal laser scanning microscope to confirm the expression of GFP. In addition, the average fluorescence intensity of each group was analyzed using ImageJ, and a relative quantitative result was obtained.
Biofilm Formation Assay
The bacteria suspensions of S. mutans UA159 in the mid-log phase (OD600=0.5) were diluted in BHIS at a volume ratio of 1:100 for the following experiments. Taking into account the effective time of DMSNs-NH2 and simulating people’s usual oral cleaning habits (gargling or brushing teeth three times during the day, no cleaning behavior at night during sleep), we designed a 12-hour group and a 24-hour group in the biofilm formation experiment. There were five subgroups in each group: (1) “UA159 + sterile deionized water” was designed as a blank control group; (2) “UA159 + pDL278” was designed to exclude the influence of the plasmid without vicR sequence on biofilm formation; (3) “UA159 + plasmid-ASvicR” was designed to explore the impact of the naked plasmid-ASvicR on biofilm formation; (4) “UA159 + DMSNs-NH2” was designed to investigate whether the nanoparticles themselves impacted biofilm formation; (5) “UA159 + DMSNs-NH2-ASvicR” was designed to verify the effect of plasmid-ASvicR loaded on DMSNs-NH2 on biofilm formation.
Concerning scanning electron microscopy (SEM), 2 mL of bacterial suspensions was added to each well of the 12-well plate containing circular glass slides and cultured at 37°C (80% N2, 10% CO2, and 10% H2) for 12 or 24 h. In the 12-hour group, sterile deionized water, naked pDL278, naked plasmid-ASvicR, DMSNs-NH2, or DMSNs-NH2-ASvicR (containing 800 ng of plasmids-ASvicR) was added every 4h. In the 24-hour group, the treatment for the first 12 hours was the same as the “12-hour group”, with no treatment for the next 12 hours. After 12 h or 24 h, the biofilms were washed with PBS three times and fixed with 2.5% glutaraldehyde in the dark for 4 h. Next, the serial concentrations of ethanol solutions were used to dehydrate biofilms in gradient. The final biofilms were observed using SEM (Inspect Hillsboro, USA).
Concerning confocal laser scanning microscopy (CLSM), bacterial solutions were added to sterile circular glass slides in 12-well plates, and 1-μM of Alexa Fluor® 647 was added to label the biofilm EPS in situ. After 12 or 24 hours co-culture, the medium was removed, and each sample was washed with sterile PBS three times. Next, 50 μL of SYTO™ 9 dye diluted by deionized water at a 1:100 ratio was added to label the bacteria for 15 min. Next, the biofilm architectures were measured by CLSM (Olympus, Japan), and Z sections at 0.8-μM intervals were used to record the thickness of the biofilms. Finally, Imaris 7.0 software (Bitplane, Switzerland) was used to reconstruct three-dimensional images of the biofilms and analyze the biomass of EPS and bacterial cells.
Quantitative Reverse Transcription Polymerase Chain Reaction
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was conducted to verify the regulatory effect of DMSNs-NH2-ASvicR on the vicR gene related to EPS synthesis. The 12-hour and 24-hour biofilms of each group (sterile deionized water, naked pDL278, naked plasmid-ASvicR, DMSNs-NH2, or DMSNs-NH2-ASvicR) were obtained as mentioned above. After being washed twice with PBS, the biofilms were harvested by a cell scraper and centrifuged (5000 rpm, 4°C, 10 min) to obtain bacterial precipitation. Total RNA was extracted using a MasterPure™ RNA purification kit and dissolved in RNase-free water. The cDNA was prepared according to the instructions of PrimeScript™ RT reagent kit with gDNA Eraser. Then, qRT-PCR was performed using LightCycler® 480 (Roche, USA), following the instructions of TB Green® Premix Ex Taq™ II. DNA gyrase subunit A (gyrA) was selected as a reference gene, and the group of “UA159 + deionized water” was used as control. Table 1 lists the corresponding primers. After amplification, each well’s quantification cycle (Cq) value was obtained, and the relative level of vicR mRNA in each group was calculated by the 2−ΔΔCt method. Western blot was also conducted (see “Western blot” section in the Supplementary materials) to further investigate the productions of VicR proteins.
 |
Table 1 Primer Information of RT-qPCR |
Cytotoxicity Assessment of DMSNs-NHs-ASvicR
The cytotoxicity of DMSNs-NH2-ASvicR was evaluated via cell counting kit-8 (CCK-8) assay and live/dead staining assay. In addition, human gingival fibroblasts (HGFs) were obtained from clinical samples and cultured in DMEM supplemented with 10% (v/v) heat-inactivated FBS at 37°C in a 5% CO2 incubator according to the literature.56
For the CCK-8 assay, the cells were seeded in a 96-well cell culture plate overnight at a density of 5000 cells/well before DMSNs-NHs-ASvicR treatment. Then, the cells were added with various concentrations of DMSNs-NH2-ASvicR (0, 32, 64, and 96 μg/mL of DMSNs-NH2 and the weight ratio of DMSNs-NH2/plasmid-ASvicR = 80). After 12 or 24 hours incubation, 10 μL of CCK-8 solution was added to each well. The 96-well plates were then incubated for 3 hours. The absorbance at 450nm was measured by SpectraMaxiD5 (Molecular Devices, USA).
For the live/dead staining assay, the cells were seeded in a 24-well cell culture plate overnight at a density of 15,000 cells/well before DMSNs-NH2-ASvicR treatment. Then, the cells were added with various concentrations of DMSNs-NH2-ASvicR (0, 32, 64, and 96 μg/mL of DMSNs-NH2 and the weight ratio of DMSNs-NH2/plasmid-ASvicR = 80). After 24 hours of incubation, the working solutions (2-μM Calcein-AM and 4.5-μM PI) and 1×assay buffer were first prepared according to the instructions. Then, the previous DMEM was removed, and 100 μL of staining solution was added to each well. The mixture was mixed and incubated for 15 min. Images of live (green fluorescence) and dead (red fluorescence) cells were obtained under a fluorescence microscope (Leica, Germany).
Animal Experiment
A rat caries model57,58 was used to study the inhibitory effects of DMSNs-NH2-ASvicR on the cariogenicity of S. mutans in vivo. Twenty-four Wistar male rats aged 21 days were provided by Dossy Experimental Animals Co., Ltd (Chengdu, China) and randomly assigned to the control group (UA159 + PBS) and experimental groups (UA159 + DMSNs-NH2, UA159 + plasmid-ASvicR, UA159 + DMSNs-NH2-ASvicR), with six rats in each group. Before the infection, 0.1% ampicillin sodium was administered for bacteriostatic treatment during 22–27 days of age. After eluting bacteriostatic treatment for one day, the saliva of those rats was collected with sterile cotton swabs and coated on the MSA mediums containing 0.2-U/mL bacitracin to detect the inhibition of endogenous S. mutans. During 28–34 days of age, each rat was infected orally once daily using 200 µL of bacterial suspension containing the UA159 strain. Meanwhile, the control group was treated with 200 µL of PBS, and the experimental groups were treated with 200 µL of DMSNs-NH2, plasmid-ASvicR, and DMSNs-NH2-ASvicR, respectively, at 28–41days of age. All the rats were fed with Keyes2000# cariogenic fodder and sterilized water containing 5% sucrose. The rats were weighed once a week and recorded daily for their health statuses. At the rats age of 56 days, they were sacrificed by CO2 asphyxiation, and their mandibles were removed and dissected into left and right halves. After washing with PBS three times, the mandibular specimens were immersed in 10% formalin solution for 48h. Then the sections were embedded and ground to be observed under a stereomicroscope to determine caries level according to a modified Keyes score.59,60 The scores for each sample were the sum of the scores for the first and second molars.
Statistical Analysis
Statistical analyses of the experimental results were performed with Prism 8 (GraphPad, USA). The Shapiro–Wilk test was used to analyze the data normality, and the Bartlett method was used to test the homogeneity of variance. If the data of each group were normally distributed and consistent with homogeneity of variance, the one-way ANOVA was used to analyze data. If the data did not exhibit normal distribution or homogeneity of variance, the Kruskal–Wallis method was used to conduct a completely random non-parametric test on the data. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 indicated statistically significant differences. All the experiments were repeated at least three times.
Results
Characterization of DMSNs-NH2
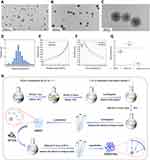
A transmission electron microscope (TEM) was used to observe the morphologies and dispersion of DMSNs-NH2, and ImageJ software was used to determine the diameters of nanoparticles. Figure 1A–D shows that DMSNs-NH2 consisted of dendrimer-like particles with center-radial pore channels. The particles were highly monodispersed, and the average particle diameter was approximately 59 nm. The information of mesopores was obtained using N2 adsorption-desorption measurements. The N2 adsorption-desorption curve of DMSNs-NH2 (Figure 1E) displayed a type-IV isotherm and type-H4 hysteresis loop, illustrating the presences of mesopores. The size distribution of mesopores (Figure 1F) was acquired from N2 adsorption isotherm via Barrett-Joyner-Halenda (BJH) method, indicating an average pore size of 8.1 nm. In addition, the Brunauer-Emmett-Teller (BET) surface areas of 449.1 m2g−1 and large total pore volumes of 0.8 cm3g−1 were also acquired. Supplementary Figure 1 and Supplementary Table 1 indicate that the DMSNs were successfully aminated. Figure 1G shows that the average potentials of the particles changed from −6.2 mV to 6.3 mV after the modification of positively charged amino groups on DMSNs. However, after loading the plasmid-ASvicR, the average potential of the particles changed to −25 mV. Figure 1H is a schematic diagram of the DMSNs-NH2 synthesis process.
Loading and Protection of Plasmid-ASvicR by DMSNs-NH2
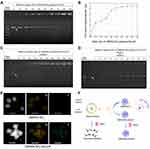
The ability of DMSNs-NH2 to load plasmid-ASvicR was confirmed by agarose gel electrophoresis and the loading curve. The bright bands in the lanes in Figure 2A represent the different forms of the plasmid-ASvicR,37 and the brightness in the wells represent the plasmids-ASvicR bound to the DMSNs-NH2 that could not run out of the wells. As the weight ratio of DMSNs-NH2/plasmid-ASvicR increased, the brightness in the well increased, while the brightness of the plasmid band in the lane decreased. Table 2 shows the loading data, and Figure 2B is a graph of plasmids loaded by DMSNs-NH2 with different qualities, and the loading trend of the graph is consistent with Figure 2A. When the mass ratio of DMSNs-NH2/plasmid-ASvicR was <80, the amount of the plasmid loaded by DMSNs-NH2 increased with an increase in the mass ratio. When the mass ratio reached 80, the plasmid was loaded 92%, and the loaded plasmid did not increase as the mass ratio continued to increase. Therefore, in the follow-up experiments, the weight ratio of 80 was used as the working ratio.
 |
Table 2 Data of Loading Curve |
The ability of DMSNs-NH2 to protect plasmid-ASvicR was confirmed by agarose gel electrophoresis. After incubation with DNase I for 15 min, DMSNs-NH2/plasmid-ASvicR complexes at different weight ratios were used for agarose gel electrophoresis. DNA bands of DMSNs-NH2/plasmid-ASvicR complexes could be observed at various weight ratios except for the group at the weight ratio of 0, indicating that the naked plasmid-ASvicR was degraded completely by DNase I without the protection of DMSNs-NH2 (Figure 2C). After treatment with heparin, the plasmid-ASvicR adsorbed on DMSNs-NH2 were released. Figure 2D shows that most of the plasmids adsorbed on DMSNs-NH2 were not degraded. The new band appearing between the two bands (a supercoiled and open-loop form of the plasmids) was a linear form that might have been caused by the vortex oscillation during the operation. EDS-mapping was performed in order to observe the distribution of plasmid-ASvicR on DMSNs-NH2 more intuitively. Figure 2E indicates that plasmid-ASvicR was distributed on the both surface and mesopores of DMSNs-NH2. However, Supplementary Figure 2 shows that the FTIR of DMSNs-NH2 and DMSNs-NH2-ASvicR had no obvious differences, which is probably because there were no chemical bonds formed between DMSNs-NH2 and plasmid-ASvicR. Figure 2F is a schematic diagram showing that DMSNs-NH2 could load and protect plasmids from degradation by DNase I.
Transformation of DMSNs-NH2-ASvicR
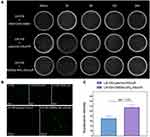
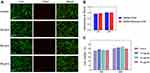
Antibiotic plate screening and fluorescence microscopy were used to determine whether DMSNs-NH2-ASvicR entered S. mutans UA159 and was expressed successfully. As shown in Figure 3A, colonies grew on the plates after being coated with the bacterial solution containing plasmid-ASvicR or DMSN-NH2-ASvicR, which had been cultured for 30 min, 3 h, and 6 h, respectively. However, there were no colonies on the plates coated with a bacterial solution cultured for 12 h and 24 h. It has been shown that UA159 and DMSNs-NH2 have no fluorescence, and pDL278 cannot make UA159 express green fluorescence. However, plasmid-ASvicR and DMSNs-NH2-ASvicR can both enter the bacteria, expressing green fluorescent protein (GFP) successfully (Figure 3B). In addition, Figure 3B and C show that the DMSNs-NH2-ASvicR group expressed a higher intensity of green fluorescence than the naked plasmid-ASvicR group (P < 0.01).
Effect of DMSNs-NH2-ASvicR on Biofilm Formation of UA159
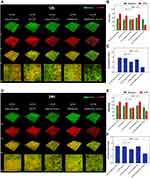
SEM and CLSM were used to evaluate the effects of DMSNs-NH2-ASvicR on biofilm formation of S. mutans UA159. Figure 4 shows the SEM images of biofilms’ morphologies of different groups. Compared with the control group, the extracellular matrix of the 12-hour biofilms in the DMSNs-NH2-ASvicR group lacked a three-dimensional structure of network crosslinking and bacteria were arranged loosely without morphological abnormality in S. mutans itself. The extracellular matrix of the plasmid-ASvicR and the DMSNs-NH2 groups was reduced to some extent, but not obviously. However, the biofilms of the pDL278 group and DMSNs-NH2 group show no significant differences from the control group. The 24-hour biofilms were generally denser than 12-hour biofilms. Even so, the biofilms of the DMSNs-NH2-ASvicR group still lacked a dense structure. It could also be seen from the 24-hour biofilms of the DMSNs-NH2-ASvicR group that nanoparticles clustered around S. mutans, and some nanoparticles even directly contacted the surface of S. mutans, which may be beneficial to plasmid transformation. The CLSM images of the 12-hour (Figure 5A) and 24-hour (Figure 5D) biofilms showed that EPS in the DMSNs-NH2-ASvicR group decreased visibly, consistent with the SEM observations. The EPS biomass and EPS/bacteria ratio in the DMSNs-NH2-ASvicR group were significantly lower than those in the control group (P < 0.0001). However, there was no significant difference in bacterial biomass (Figure 5B, C, E and F). Meanwhile, EPS biomass and EPS/bacteria ratio in the plasmid-ASvicR group also decreased but not as significantly as in the DMSNs-NH2-ASvicR group.
Regulation of vicR Gene Expression in Streptococcus mutans
RT-qPCR was performed on mRNA extracted from 12-hour and 24-hour biofilms to demonstrate the regulatory effect of DMSNs-NH2 on the target gene vicR. As shown in Figure 6A, the pDL278 and DMSNs-NH2 groups show no effect on the expression of vicR. Nevertheless, the relative quantity of vicR was down-regulated by 0.74 times (P < 0.0001) in the 12-hour biofilms and 0.83 times (P < 0.0001) in 24-hour biofilms of the DMSNs-NH2-ASvicR group. In addition, the level of vicR was also down-regulated by 0.91 times (P < 0.01) in the 12-hour biofilms of the plasmid-ASvicR group, with no significant difference in 24-hour biofilms compared to the control group. Supplementary Figure 3 is the result of Western blot of 24-hour biofilms, which is consistent with result of RT-qPCR. Figure 6B displays the mechanism of DMSNs-NH2-ASvicR regulating biofilm formation of biofilms and the expression of vicR in S. mutans.
Cytotoxicity Assessment of DMSNs-NH2-ASvicR
In vitro cytotoxicity of DMSNs-NH2-ASvicR was evaluated by CCK-8 assay and live/dead staining assays. The fluorescence micrograph of live/dead staining assay showed that most HGFs were viable (green fluorescence) without morphological abnormality after being treated with different concentrations of DMSNs-NH2-ASvicR, with very few dead cells (red fluorescence) similar to the control group (Figure 7A). As shown in Figure 7B, the influence of DMSNs-NH2-ASvicR on absorbance at 450nm was excluded. DMSNs-NH2-ASvicR had no adverse effects on the viability of HGFs after 12 and 24 hours of incubation, even at a concentration of 96 μg/μL (Figure 7C).
DMSNs-NH2-ASvicR Decreased the Cariogenicity of S. mutans in vivo
A rat caries model was used to investigate the effects of DMSNs-NH2-ASvicR on the cariogenicity of S. mutans in vivo. Figure 8 presents the stereomicroscopic images and Keys scoring results of rat mandibular molars. “E” scored 1 point, indicating that caries only involved enamel. “Ds” scored 2 points, meaning that caries involved no more than 1/4 of the dentin thickness. “Dm” scored 3 points, implying that caries involved 1/4 to 3/4 of dentin thickness. “Dx” scored 4 points, indicating that caries involved more than 3/4 of dentin thickness. The DMSNs-NH2-ASvicR group had no apparent caries or only a few shallow carious cavities, and the total Keys score was significantly lower than the control group (P < 0.01, Figure 8B). There was also a decrease in caries in the plasmid-ASvicR group, however, with no statistical significance.
 |
Figure 8 (A) Stereoscopic microscopy of pit fissure caries lesions on mandibular molars of rats. (B) Total Keys scores of caries on the first and second mandibular molars of rats. **P < 0.01, n = 10. |
Discussion
It is well established that larger pore volume and surface area of nanoparticles provide more spaces for delivering biomacromolecules.38,61 Studies have shown that mesoporous silica nanoparticles with smaller sizes could induce higher cell uptake.54,62,63 In the present study, the bacteria were much smaller than cells; thus, nanoparticles with sufficiently small sizes were required. Therefore, we synthesized DMSNs-NH2, with morphology similar to that reported in the literatures.42,50,51 In addition, the particles were smaller in diameter (~59 nm), because we reduced the reaction time according to the literature.50 The DMSNs-NH2 has a large pore size of 8.1 nm, a surface area of 449.1 m2g−1, and a pore volume of 0.8 cm3g−1, which are beneficial for loading and protecting plasmid DNA and increasing the delivery efficiency.38,54,64 It is worth mentioning that according to literature, mesoporous silica nanoparticles with a hollow architecture showed higher loading efficiency than those with the same mesoporous morphology but no hollow architecture.44,65 This reminds us that we can add the hollow architecture based on DMSNs-NH2 in future research, which might be more helpful in improving the efficiency of loading and transferring the plasmid.
The positive potential is essential to ensure the ability of DMSNs-NH2 to encapsulate and deliver negatively charged plasmid-ASvicR.52 However, the average zeta potential of DMSNs-NH2 synthesized in the present study was only 6.3 mV, which is lower than that reported in the literature.37,54,66 Therefore, the efficiency of DMSNs-NH2 loading plasmid-ASvicR was not high enough; thus, a higher mass ratio (DMSNs-NH2/plasmid-ASvicR=80) is required to load plasmid-ASvicR completely. Hence, it is one of the goals in our future work to improve the amination method to increase loading efficiency. The ability of DMSNs-NH2 to load and protect plasmid-ASvicR has also been demonstrated. When DMSNs-NH2 was mixed with plasmid-ASvicR by mass at a ratio of 80, the plasmid-ASvicR could be loaded 92% and most plasmids could be protected from degradation.
It has been reported that bacteria can take up extracellular free DNA naturally.67–69 The transformation assays in the present study indicated that the naked plasmid-ASvicR could enter S. mutans UA159. Furthermore, the plasmids loaded on the DMSNs-NH2 could successfully enter S. mutans, express related genes, and exhibit higher transformation efficiency than naked plasmids. The possible reasons are as follows: (1) DMSNs-NH2 increased the stability of the plasmids in the bacterial solution;54 (2) DMSNs-NH2 loaded with plasmid-ASvicR was prone to aggregation and adsorption by bacteria, which increased the chance of plasmid contacting the bacteria, promoting the entry of plasmids into the bacteria. However, the exact mechanism remains unknown, which is one of the limitations of the present study, necessitating further investigations. There were no colonies in the 12-hour and 24-hour group, perhaps because ASvicR entered S. mutans and interfered with the expression of vicR, which has been reported to be critical for viability,8,19 thereby reducing their survival advantage. These findings imply that DMSNs-NH2-ASvicR can enter the bacteria and affect the expression of related genes, but the effective time is short. That is why we treated the bacteria with samples every 4 hours in the subsequent biofilm formation assay.
Antibiotics have been used to fight bacterial infections for several decades. However, most antibiotics used to treat bacterial infections have several side effects and induce drug resistance.70–72 Therefore, it is necessary to develop highly effective antibacterial agents to address this threatening resistance crisis. The results of biofilm formation assays and RT-qPCR indicate that pDL278 and DMSNs-NH2 did not impact the cariogenicity of S. mutans compared to the blank control. However, DMSNs-NH2-ASvicR could down-regulate the expression of vicR and reduce the productions of EPS by S. mutans, affecting the morphology and structure of the biofilm. Interestingly, although DMSNs-NH2-ASvicR affected the biofilms, it did not change the morphology and quantity of bacteria itself, which might be conducive to maintaining the homeostasis of the microecological flora. The CLSM results showed that EPS of the plasmid-ASvicR group was also decreased, with no significant changes in the biofilm structure under SEM. One possible explanation was that although the naked plasmid-ASvicR could enter S. mutans and affect the synthesis of EPS, the transformation efficiency of the naked plasmid-ASvicR entering S. mutans was too limited to cause significant changes in biofilm morphology. Both DMSNs-NH2-ASvicR and plasmid-ASvicR showed lower regulatory effects on the biofilm formation after 24 h, indicating that the frequency of treatment should be increased or a controlled release system should be used to achieve long-term ideal biofilm inhibitory effects.
For biomedical applications, the delivery system should possess functional performance and excellent biocompatibility.73 The cytotoxicity experiments indicated that DMSNs-NH2-ASvicR had no cytotoxicity, providing a basis for the subsequent application in vivo. The results of Keys scores showed that DMSNs-NH2-ASvicR could reduce caries incidence in rats significantly, indicating that DMSNs-NH2 could effectively protect the plasmid-ASvicR from degradation and facilitate its entry into the bacteria in the rat’s oral cavity. There was also a decrease in caries with no statistical significance in the plasmid-ASvicR group compared with the blank control. It was speculated that some plasmid-ASvicR could enter the bacteria before the plasmids were degraded by nuclease in the rat’s mouth. Preliminarily, the results indicated the regulatory effect of DMSNs-NH2-ASvicR on the cariogenicity of S. mutans in vivo, laying a foundation for further studies. Nevertheless, there are some limitations in the animal experiment, necessitating more concentration gradients and longer research time.
Conclusion
DMSNs-NH2 synthesized in the present study has been verified to possess the capacity of delivering plasmid-ASvicR and protecting them from degradation. With the protection of DMSNs-NH2, plasmid-ASvicR could enter S. mutans and down-regulate the expression of the vicR gene, reducing the synthesis of EPS and affecting the formation of biofilm. In vivo experiments confirmed that DMSNS-NH2-ASvicR could reduce the cariogenicity of S. mutans preliminarily. Nevertheless, the specific mechanisms of DMSNs-NH2-ASvicR in regulating the S. mutans should be further explored. It is expected that DMSNs-NH2-ASvicR can undergo clinical transformation to become a substance for dental caries prevention in the future.
Ethics Statement
The human gingival fibroblasts (HGFs) used in the study were obtained from clinical samples, which were carried out in accordance with the recommendations of the Institutional Ethics Committee of West China Hospital of Stomatology. The involved subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Institutional Ethics Committee of West China Hospital of Stomatology (WCHSIRB-D-2020-075-R1).
The experiments involving rats were carried out in accordance with the Chinese State Key Laboratory of Oral Diseases guidelines for animal welfare and were approved by the Institutional Ethics Committee of West China Hospital of Stomatology (WCHSIRB-D-2021-336).
Acknowledgments
This study was supported by the National Natural Science Foundation of China (82170948, 51872190), Interdisciplinary Innovation Projects of Sichuan University (RD-03-202002), Sichuan Provincial Natural Science Foundation of China (2020YFH0010), Basic and Applied Research Project of West China Hospital of Stomatology (RD-02-202001), and Sichuan University new century higher education teaching reform project (the ninth phase) research project (SCU9361).
Author Contributions
All authors made significant contributions to conception, study design, execution, acquisition of data, or analysis and interpretation of data. All authors took part in drafting the article, revising substantially, or reviewing the article critically. All authors agreed to submit to the current journal, gave final approval of the version to be published, and agreed to be accountable for the contents of the article.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bernabe E, Marcenes W, Hernandez CR, et al. Global, regional, and national levels and trends in burden of oral conditions from 1990 to 2017: a systematic analysis for the global burden of disease 2017 study. J Dent Res. 2020;99(4):362–373. doi:10.1177/0022034520908533
2. Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B, Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33(4):499–515. doi:10.1007/s10096-013-1993-7
3. Lemos JA, Palmer SR, Zeng L, et al. The biology of Streptococcus mutans. Microbiol Spectr. 2019;7(1):1–18. doi:10.1128/microbiolspec.gpp3-0051-2018
4. Koo H, Falsetta ML, Klein MI. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent Res. 2013;92(12):1065–1073. doi:10.1177/0022034513504218
5. Bowen WH, Koo H. Biology of streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86. doi:10.1159/000324598
6. Bowen WH. Dental caries – not just holes in teeth! A perspective. Mol Oral Microbiol. 2016;31(3):228–233. doi:10.1111/omi.12132
7. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
8. Senadheera MD, Guggenheim B, Spatafora GA, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187(12):4064–4076. doi:10.1128/JB.187.12.4064
9. Ajdić D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99(22):14434–14439. doi:10.1073/pnas.172501299
10. Deng DM, Liu MJ, Ten Cate JM, Crielaard W. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res. 2007;86(7):606–610. doi:10.1177/154405910708600705
11. Lei L, Long L, Yang X, et al. The VicRK two-component system regulates Streptococcus mutans virulence. Curr Issues Mol Biol. 2019;32:167–200. doi:10.21775/cimb.032.167
12. André G, Even S, Putzer H, et al. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36(18):5955–5969. doi:10.1093/nar/gkn601
13. Georg J, Vob B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5(1):305. doi:10.1038/msb.2009.63
14. Dornenburg JE, DeVita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010;1(1):1–4. doi:10.1128/mBio.00024-10
15. Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–355.
16. Sully EK, Geller BL. Antisense antimicrobial therapeutics. Curr Opin Microbiol. 2016;33:47–55. doi:10.1016/j.mib.2016.05.017
17. Hegarty JP, Stewart DB. Advances in therapeutic bacterial antisense biotechnology. Appl Microbiol Biotechnol. 2018;102(3):1055–1065. doi:10.1007/s00253-017-8671-0
18. Lei L, Stipp RN, Chen T, Wu SZ, Hu T, Duncan MJ. Activity of Streptococcus mutans VicR is modulated by antisense RNA. J Dent Res. 2018;97(13):1477–1484. doi:10.1177/0022034518781765
19. Lei L, Zhang B, Mao M, et al. Carbohydrate metabolism regulated by antisense vicR RNA in cariogenicity. J Dent Res. 2020;99(2):204–213. doi:10.1177/0022034519890570
20. Cha W, Fan R, Miao Y, et al. Mesoporous silica nanoparticles as carriers for intracellular delivery of nucleic acids and subsequent therapeutic applications. Molecules. 2017;22(5):782. doi:10.3390/molecules22050782
21. Shen H, Huang X, Min J, et al. Nanoparticle delivery systems for DNA/RNA and their potential applications in nanomedicine. Curr Top Med Chem. 2019;19(27):2507–2523. doi:10.2174/1568026619666191024170212
22. Ginocchio VM, Ferla R, Auricchio A, Brunetti-Pierri N. Current status on clinical development of adeno-associated virus-mediated liver-directed gene therapy for inborn errors of metabolism. Hum Gene Ther. 2019;30(10):1204–1210. doi:10.1089/hum.2019.151
23. Buck J, Grossen P, Cullis PR, Huwyler J, Witzigmann D. Lipid-based DNA therapeutics: hallmarks of non-viral gene delivery. ACS Nano. 2019;13(4):3754–3782. doi:10.1021/acsnano.8b07858
24. Lehto T, Ezzat K, Wood MJA, Andaloussi SEL. Peptides for nucleic acid delivery. Adv Drug Deliv Rev. 2016;106:172–182. doi:10.1016/j.addr.2016.06.008
25. Halman JR, Kim KT, Gwak SJ, et al. A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine. 2020;23:102094. doi:10.1016/j.nano.2019.102094
26. Loh XJ, Lee TC, Dou Q, Deen GR. Utilising inorganic nanocarriers for gene delivery. Biomater Sci. 2016;4(1):70–86. doi:10.1039/c5bm00277j
27. Zhou Y, Quan G, Wu Q, et al. Mesoporous silica nanoparticles for drug and gene delivery. Acta Pharm Sin B. 2018;8(2):165–177. doi:10.1016/j.apsb.2018.01.007
28. Neshat SY, Tzeng SY, Green JJ. Gene delivery for immunoengineering. Curr Opin Biotechnol. 2020;66:1–10. doi:10.1016/j.copbio.2020.05.008
29. Bai Z, Wei J, Yu C, et al. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J Mater Chem B. 2019;7(8):1209–1225. doi:10.1039/c8tb02946f
30. Xue XY, Mao XG, Zhou Y, et al. Advances in the delivery of antisense oligonucleotides for combating bacterial infectious diseases. Nanomedicine. 2018;14(3):745–758. doi:10.1016/j.nano.2017.12.026
31. Tarn D, Ashley CE, Xue M, Carnes E, Zink JI, Brinker J. Mesoporous silica nanoparticle nanocarriers: biofunctionality and biocompatibility. Acc Chem Res. 2013;46(3):792–801. doi:10.1021/ar3000986
32. Narayan R, Nayak UY, Raichur AM, Garg S. Mesoporous silica nanoparticles: a comprehensive review on synthesis and recent advances. Pharmaceutics. 2018;10(3):1–49. doi:10.3390/pharmaceutics10030118
33. Ghaferi M, Koohi Moftakhari Esfahani M, Raza A, Al Harthi S, Ebrahimi Shahmabadi H, Alavi SE. Mesoporous silica nanoparticles: synthesis methods and their therapeutic use-recent advances. J Drug Target. 2021;29(2):131–154. doi:10.1080/1061186X.2020.1812614
34. Kankala RK, Liu CG, Yang DY, Wang S-B, Chen AZ. Ultrasmall platinum nanoparticles enable deep tumor penetration and synergistic therapeutic abilities through free radical species-assisted catalysis to combat cancer multidrug resistance. Chem Eng J. 2020;383:123138. doi:10.1016/j.nano.2017.12.026
35. Kesse S, Boakye-Yiadom K, Ochete BO, et al. Mesoporous silica nanomaterials: versatile nanocarriers for cancer theranostics and drug and gene delivery. Pharmaceutics. 2019;11(2):1–26. doi:10.3390/pharmaceutics11020077
36. Gao F, Botella P, Corma A, Blesa J, Dong L. Monodispersed mesoporous silica nanoparticles with very large pores for enhanced adsorption and release of DNA. J Phys Chem B. 2009;113(6):1796–1804. doi:10.1021/jp807956r
37. Kim MH, Na HK, Kim YK, et al. Facile synthesis of monodispersed mesoporous silica nanoparticles with ultralarge pores and their application in gene delivery. ACS Nano. 2011;5(5):3568–3576. doi:10.1021/nn103130q
38. Wu M, Meng Q, Chen Y, et al. Large-pore ultrasmall mesoporous organosilica nanoparticles: micelle/precursor co-templating assembly and nuclear-targeted gene delivery. Adv Mater. 2015;27(2):215–222. doi:10.1002/adma.201404256
39. Kankala RK, Han YH, Na J, et al. Nanoarchitectured structure and surface biofunctionality of mesoporous silica nanoparticles. Adv Mater. 2020;32(23):23. doi:10.1002/adma.201907035
40. Wang Y, Zhang B, Ding X, Du X. Dendritic mesoporous organosilica nanoparticles (DMONs): chemical composition, structural architecture, and promising applications. Nano Today. 2021;39:101231. doi:10.1016/j.nantod.2021.101231
41. Du X, Li X, Huang H, He J, Zhang X. Dendrimer-like hybrid particles with tunable hierarchical pores. Nanoscale. 2015;7(14):6173–6184. doi:10.1039/c5nr00640f
42. Shen D, Chen L, Yang J, et al. Ultradispersed palladium nanoparticles in three-dimensional dendritic mesoporous silica nanospheres: toward active and stable heterogeneous catalysts. ACS Appl Mater Interfaces. 2015;7(31):17450–17459. doi:10.1021/acsami.5b04992
43. Du X, Zhang qiao S. Dendritic silica particles with center-radial pore channels: promising platforms for catalysis and biomedical applications. Small. 2015;11(4):392–413. doi:10.1002/smll.201401201
44. Yang Y, Bernardi S, Song H, et al. Anion assisted synthesis of large pore hollow dendritic mesoporous organosilica nanoparticles: understanding the composition gradient. Chem Mater. 2016;28(3):704–707. doi:10.1021/acs.chemmater.5b03963
45. Chang J-H, Tsai P-H, Chen W, Chiou S-H, Mou C-Y. Dual delivery of siRNA and plasmid DNA using mesoporous silica nanoparticles to differentiate induced pluripotent stem cells into dopaminergic neurons. J Mater Chem B. 2017;5(16):3012–3023. doi:10.1039/c7tb00351j
46. Wang Y, Ding X, Chen Y, et al. Antibiotic-loaded, silver core-embedded mesoporous silica nanovehicles as a synergistic antibacterial agent for the treatment of drug-resistant infections. Biomaterials. 2016;101:207–216. doi:10.1016/j.biomaterials.2016.06.004
47. Lu MM, Wang QJ, Chang ZM, et al. Synergistic bactericidal activity of chlorhexidineloaded, silver-decorated mesoporous silica nanoparticles. Int J Nanomedicine. 2017;12:3577–3589. doi:10.2147/IJN.S133846
48. Sun J, Fan Y, Zhang P, et al. Self-enriched mesoporous silica nanoparticle composite membrane with remarkable photodynamic antimicrobial performances. J Colloid Interface Sci. 2020;559:197–205. doi:10.1016/j.jcis.2019.10.021
49. Castillo RR, Vallet-regí M. Recent advances toward the use of mesoporous silica nanoparticles for the treatment of bacterial infections. Int J Nanomedicine. 2021;Volume 16:4409–4430. doi:10.2147/IJN.S273064
50. Shen D, Yang J, Li X, et al. Biphase stratification approach to three-dimensional dendritic biodegradable mesoporous silica nanospheres. Nano Lett. 2014;14(2):923–932. doi:10.1021/nl404316v
51. Hong X, Zhong X, Du G, et al. The pore size of mesoporous silica nanoparticles regulates their antigen delivery efficiency. Sci Adv. 2020;6(25). doi:10.1126/sciadv.aaz4462
52. Keasberry NA, Yapp CW, Idris A. Mesoporous silica nanoparticles as a carrier platform for intracellular delivery of nucleic acids. Biochem. 2017;82(6):655–662. doi:10.1134/S0006297917060025
53. Zarei H, Kazemi Oskuee R, Hanafi-Bojd MY, Gholami L, Ansari L, Malaekeh-Nikouei B. Enhanced gene delivery by polyethyleneimine coated mesoporous silica nanoparticles. Pharm Dev Technol. 2019;24(1):127–132. doi:10.1080/10837450.2018.1431930
54. Tao C, Zhu Y, Xu Y, Zhu M, Morita H, Hanagata N. Mesoporous silica nanoparticles for enhancing the delivery efficiency of immunostimulatory DNA drugs. Dalton Trans. 2014;43(13):5142–5150. doi:10.1039/c3dt53433b
55. Bottaro LB. Mesoporous silica nanoparticles facilitate delivery of siRNA to shutdown signaling pathways in mammalian cells. small. 2011;23(1):1–7. doi:10.1002/smll.200901966.Mesoporous
56. Cheng R, Liu W, Zhang R, Feng Y, Bhowmick NA, Hu T. Porphyromonas gingivalis-derived lipopolysaccharide combines hypoxia to induce caspase-1 activation in periodontitis. Front Cell Infect Microbiol. 2017;7:1–9. doi:10.3389/fcimb.2017.00474
57. Lei L, Yang Y, Mao M, et al. Modulation of biofilm exopolysaccharides by the Streptococcus mutans vicX gene. Front Microbiol. 2015;6:1–14. doi:10.3389/fmicb.2015.01432
58. Mao MY, Yang YM, Li KZ, et al. The rnc gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via microRNA-size small RNAs in Streptococcus mutans. Front Microbiol. 2016;7:1–11. doi:10.3389/fmicb.2016.00687
59. Keyes PH. Dental Caries in the molar teeth of rats: I. Distribution of lesions induced by high-carbohydrate low-fat diets. J Dent Res. 1958;37(6):1077–1087. doi:10.1177/00220345580370060801
60. Su LK, Yu F, Li ZF, Zeng C, Xu QA, Fan MW. Intranasal co-delivery of IL-6 gene enhances the immunogenicity of anti-caries DNA vaccine. Acta Pharmacol Sin. 2014;35(5):592–598. doi:10.1038/aps.2013.184
61. Li X, Wong CH, Ng TW, Zhang CF, Leung KCF, Jin L. The spherical nanoparticle-encapsulated chlorhexidine enhances anti-biofilm efficiency through an effective releasing mode and close microbial interactions. Int J Nanomedicine. 2016;11:2471–2480. doi:10.2147/IJN.S105681
62. Lu F, Wu SH, Hung Y, Mou CY. Size effect on cell uptake in well-suspended, uniform mesoporous silica nanoparticles. Small. 2009;5(12):1408–1413. doi:10.1002/smll.200900005
63. Gan Q, Dai D, Yuan Y, et al. Effect of size on the cellular endocytosis and controlled release of mesoporous silica nanoparticles for intracellular delivery. Biomed Microdevices. 2012;14(2):259–270. doi:10.1007/s10544-011-9604-9
64. Guo Z, Wu L, Wang Y, et al. Design of dendritic large-pore mesoporous silica nanoparticles with controlled structure and formation mechanism in dual-templating strategy. ACS Appl Mater Interfaces. 2020;12(16):18823–18832. doi:10.1021/acsami.0c00596
65. Lee JY, Kim MK, Nguyen TL, Kim J. Hollow mesoporous silica nanoparticles with extra-large mesopores for enhanced cancer vaccine. ACS Appl Mater Interfaces. 2020;12(31):34658–34666. doi:10.1021/acsami.0c09484
66. Du X, Shi B, Liang J, Bi J, Dai S, Qiao SZ. Developing functionalized dendrimer-like silica nanoparticles with hierarchical pores as advanced delivery nanocarriers. Adv Mater. 2013;25(41):5981–5985. doi:10.1002/adma.201302189
67. Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi:10.1038/nrmicro844
68. Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12(3):181–196. doi:10.1038/nrmicro3199
69. Hasegawa H, Suzuki E, Maeda S. Horizontal plasmid transfer by transformation in Escherichia coli: environmental factors and possible mechanisms. Front Microbiol. 2018;9:1–6. doi:10.3389/fmicb.2018.02365
70. Jayakumar J, Kumar VA, Biswas L, Biswas R. Therapeutic applications of lysostaphin against Staphylococcus aureus. J Appl Microbiol. 2020;1–11. doi:10.1111/jam.14985
71. Jiang YL, Qiu W, Zhou XD, et al. Quaternary ammonium-induced multidrug tolerant streptococcus mutans persisters elevate cariogenic virulence in vitro. Int J Oral Sci. 2017;9(12):e7–8. doi:10.1038/ijos.2017.46
72. Song X, Liu P, Liu X, et al. Dealing with MDR bacteria and biofilm in the post-antibiotic era: application of antimicrobial peptides-based nano-formulation. Mater Sci Eng C. 2021;128:112318. doi:10.1016/j.msec.2021.112318
73. Tang F, Li L, Chen D. Mesoporous silica nanoparticles: synthesis, biocompatibility and drug delivery. Adv Mater. 2012;24(12):1504–1534. doi:10.1002/adma.201104763
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.