Back to Journals » Infection and Drug Resistance » Volume 15
Antimicrobial Resistance Rates and Surveillance in Sub-Saharan Africa: Where Are We Now?
Authors Kariuki S, Kering K , Wairimu C, Onsare R, Mbae C
Received 15 March 2022
Accepted for publication 16 June 2022
Published 7 July 2022 Volume 2022:15 Pages 3589—3609
DOI https://doi.org/10.2147/IDR.S342753
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Samuel Kariuki, Kelvin Kering, Celestine Wairimu, Robert Onsare, Cecilia Mbae
Centre for Microbiology Research, Kenya Medical Research Institute, Nairobi, Kenya
Correspondence: Samuel Kariuki, Centre for Microbiology Research, Kenya Medical Research Institute, Nairobi, Kenya, Email [email protected]
Introduction: Although antimicrobials have traditionally been used to treat infections and improve health outcomes, resistance to commonly used antimicrobials has posed a major challenge. An estimated 700,000 deaths occur globally every year as a result of infections caused by antimicrobial-resistant pathogens. Antimicrobial resistance (AMR) also contributes directly to the decline in the global economy. In 2019, sub-Saharan Africa (SSA) had the highest mortality rate (23.5 deaths per 100,000) attributable to AMR compared to other regions.
Methods: We searched PubMed for articles relevant to AMR in pathogens in the WHO-GLASS list and in other infections of local importance in SSA. In this review, we focused on AMR rates and surveillance of AMR for these priority pathogens and some of the most encountered pathogens of public health significance. In addition, we reviewed the implementation of national action plans to mitigate against AMR in countries in SSA.
Results and Discussion: The SSA region is disproportionately affected by AMR, in part owing to the prevailing high levels of poverty, which result in a high burden of infectious diseases, poor regulation of antimicrobial use, and a lack of alternatives to ineffective antimicrobials. The global action plan as a strategy for prevention and combating AMR has been adopted by most countries, but fewer countries are able to fully implement country-specific action plans, and several challenges exist in many settings.
Conclusion: A concerted One Health approach will be required to ramp up implementation of action plans in the region. In addition to AMR surveillance, effective implementation of infection prevention and control, water, sanitation, and hygiene, and antimicrobial stewardship programs will be key cost-effective strategies in helping to tackle AMR.
Keywords: antimicrobial resistance, surveillance, sub-Saharan Africa, GAP, NAP
Background
Antimicrobial resistance (AMR) is a huge challenge to public health and directly impacts economic growth globally, with developing countries in Africa bearing the biggest burden of the negative effects of AMR.1,2 AMR is the ability of microorganisms to persist or grow in the presence of drugs designed to inhibit or kill them. This results in therapeutic failure, which negatively impacts the global control and management of infectious diseases. Infections caused by antimicrobial-resistant pathogens are reported to result in approximately 700,000 deaths globally every year.3 Furthermore, if no intervention strategies are initiated, it is estimated that by 2050, mortalities attributed to AMR will have increased to 10 million annually,4 with Africa and South Asia bearing the highest burden of deaths.5
Impact of Antimicrobial Resistance
The impact of AMR can be classified as either clinical or economic.6 The clinical effects of AMR include severe infections, increased morbidity, treatment failure, and mortality.7 In 2019, an estimated 4.95 million deaths were associated with AMR globally, with 1.27 million deaths being attributed to AMR.8 Lower respiratory and other chest infections, bloodstream and intra-abdominal infections accounted for 78.8% of mortalities attributed to antimicrobial-resistant infections. Compared to other Global Burden of Disease study regions, sub-Saharan Africa (SSA) and South Asia had the highest mortalities (23.5 deaths per 100,000 and 21.5 deaths per 100,000, respectively) attributable to AMR. Globally, Western SSA had the highest death rate attributable to AMR, 27.3 deaths per 100,000, followed by Eastern, Central, and Southern SSA (Table 1).8 The high death rate attributable to AMR in SSA is a plausible effect of ineffective water, sanitation, and hygiene (WASH) infrastructure,9,10 inefficient infection prevention and control, poor regulation of antimicrobial use, and constrained healthcare systems. In neonatal sepsis, 31% of mortalities (690,000) reported were plausibly attributed to antimicrobial-resistant infections.11
 |
Table 1 Death Rates Involving Infection, Associated with and Attributable to AMR, in SSA in 2019 |
The economic effects of AMR comprise increased length of hospitalization, higher healthcare costs,7,12 and decreased labor supply and labor efficiency.13 The global economic cost associated with AMR is estimated to be US$ 100 trillion by 2050 if the current trend persists.14 Currently, the economic cost associated with AMR in Europe is estimated to be more than €2.0 million, while in the USA the cost is approximately $55 billion.15 According to projections by RAND Europe, the economic loss due to AMR would be the highest in the SSA region if no intervention were implemented.16
Role of Antimicrobial Use in Increasing Rates of AMR
AMR essentially occurs as the result of the acquisition, expression, and transfer of resistance genes in pathogens. Pathogens acquire resistance genes through natural selection when exposed to antimicrobial pressure.16,17 Indeed, the rapid emergence of antimicrobial-resistant pathogens is often accelerated by the inappropriate use and overuse of antimicrobial agents, especially antibiotics, in humans, animals, and agriculture/the environment.18,19 While antimicrobials are often used to improve treatment outcomes, inappropriate and/or unnecessary consumption of antimicrobials by humans is a huge contributor to the emergence of AMR. Inappropriate usage/consumption of antimicrobials in humans occurs through inappropriate prescription,20 needlessly lengthy courses of treatment,21 self-medication,22–25 incomplete use of prescribed antimicrobials, and irregular use of reserve antimicrobials.
A study published in 2021 reported that there was a 46% increase in the global consumption of antimicrobials between 2000 and 2018.26 The study found that the consumption rate increased from 9.8 (9.2–10.5) daily defined doses (DDD) to 14.3 (13.2–15.6) DDD per 1000 population per day. According to the World Health Organization (WHO) report on surveillance of antimicrobial consumption, in 65 countries, consumption ranged from 4.4 to 64.4 DDD per 1000 population per day.27 In the four SSA countries assessed, consumption ranged from 4.4 DDD in Burundi to 27.29 DDD per 1000 population per day in Tanzania.
Owing to the global increase in human population, demand for food, both plant and animal products, has increased. Nearly three-quarters of all antimicrobials sold globally are used in food-producing animals.28 In food-producing animals, antimicrobials are mainly used for the prevention and treatment of disease.19,29,30 Although the use of antimicrobials is important in ensuring the health of the animals, prolonged use for non-therapeutic purposes (eg prophylaxis, growth promotion), and often at sub-therapeutic levels, contributes to the emergence and persistence of AMR.29
The use of antimicrobials in animals contributes to AMR in both animals and humans. Study reports have shown that between 2015 and 2019, 3558–4279 tons of antimicrobials were used in animal production in 13–27 African countries.31 Globally, the largest AMR hotspots in animal health are China and India. However, Brazil and Kenya have also been identified as important hotspots for the emergence of AMR attributed to use of antimicrobials in animals.29
It has been shown that the reduction of inappropriate antimicrobial use directly contributes to the decrease in the development of AMR; however, this alone would not be adequate to deal with the global AMR challenge.32 Consequently, there is a need to control the high rate of emergence and spread of antimicrobial-resistant strains and genes in the environment, animals, and human population (the One Health approach).
Methodology
In this review, “sub-Saharan Africa” refers to the countries that constitute the WHO- Africa region. The SSA region was then divided into four sub-regions (Western, Eastern, Central, and Southern). Relevant English literature (original articles and reviews) from the four sub-regions was retrieved from PubMed and Google Scholar and considered for review. The literature search was based on several search terms: antimicrobial resistance, multi-drug resistance, sub-Saharan Africa, surveillance, global action plan, national action plan, and One Health approach. The pathogens discussed were selected on the basis of being priority pathogens for surveillance and reporting to the WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS),33,34 with Vibrio cholerae being included as an important neglected tropical disease and with emerging multidrug-resistant strains from the region. The inclusion criteria also included articles on the selected pathogens from SSA countries, articles reporting on antibiotic resistance rates and genes, and articles published between the years 2000 and 2022. These criteria were inclusive of both monomicrobial and polymicrobial studies. The abstracts of all articles that met the above criteria were reviewed. Where insufficient detail was provided, the entire article was reviewed before determining whether to include it or not. Exclusion criteria were articles on non-WHO-GLASS pathogens, articles from beyond the SSA region, conference abstracts, and intervention trials. Duplicate references and publications reporting the same data in different journals were excluded.
Results and Discussion
Antimicrobial Resistance Rates in Sub-Saharan Africa
AMR is a challenge in both developed and developing countries. However, AMR prevalence varies considerably geographically in different continents, regions, and countries.35 In SSA, different pathogens were found to have varying rates of resistance to different antimicrobial agents and these may be different from region to region. This review focused on AMR among key enteric infectious diseases of public health significance in the SSA region.
Diarrheagenic Escherichia coli
Diarrheagenic Escherichia coli, especially enteropathogenic E. coli (EPEC) and enterotoxigenic E. coli (ETEC), accounts for 14.26 million (5.63–30.90 million) and 222.64 million (144.95–322.85 million) episodes each year, respectively. According to the 2016 Global Burden of Disease (GBD) report, cumulatively these two pathotypes result in 63,523 deaths among all ages annually, throughout the world.36 Other important E. coli pathotypes have been implicated, namely: enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), enteroinvasive E. coli (EIEC), and diffusely adhering E. coli (DAEC), which are important in specific regions globally.37 In SSA, the precise burden associated with diarrheagenic E. coli is not known; however, prevalence rates ranging from 14.7% to 82% have been reported in Nigeria,37 Gabon,38 Burkina Faso,39 Kenya,40 South Africa,41 and Tanzania.42 Children under the age of five have the highest risk of infection in the region. Transmission is via the fecal route, either directly through person-to-person contact or indirectly through contaminated food and water. Primarily, the diagnosis of diarrheagenic E. coli in patients and populations is through physical examination, signs and symptoms, and laboratory diagnosis – which is not routinely performed in primary care facilities owing to inadequate infrastructure. Confirmatory tests for diarrheagenic E. coli pathotypes are carried out in tertiary and research laboratories, where molecular methods, both culture-based and culture-independent assays, are utilized.
The standard treatment for diarrheagenic E. coli is oral rehydration, and antimicrobials are only recommended in severe cases. Over the years, antimicrobial-resistant diarrheagenic E. coli has emerged and varying resistance rates have been reported (Table 2). Resistance genes harbored in a variety of mobile genetic elements have been implicated: blaTEM, blaSHV, blaCTX-M-15, blaOXA-2, blaAmpC, blaOXA-181, blaNDM, catA, tetA, sulI, II & III, dfrA, aac(6’)lb, aada1, qnrS1, and rmtA.41,43–49 Of concern are carbapenemase-encoding genes, which have been observed among diarrheagenic E. coli isolates in several countries in the region. For instance, in Ghana47 and Burkina Faso,46 ST410 diarrheagenic E. coli isolates were reported to have the blaOXA-181 gene (a variant of the class D blaOXA-48), which was contained in an IncX3 plasmid. Another diarrheagenic E. coli isolate in Ghana, of the sequence type 940, was also demonstrated to have blaOXA-181 but in an IncFIC (FII) plasmid. In addition, it harbored the qnrS1 gene, which confers resistance to fluoroquinolones in an IS26 transposon.47 An ST410 with the blaNDM-1 gene contained in an IncHI1, as well as Tn125 transposon, and an ISSm-1-like insertion sequence belonging to the novel IS110 family has also been reported, further raising concerns regarding the effectiveness of carbapenems in treatment.45
 |
Table 2 Antibiotic Resistance Rates of Clinical Diarrheagenic E. coli (DEC) in SSA |
Currently, no licensed vaccine is available for diarrheagenic E. coli, but several candidates are in different stages of development.51 Control measures are needed to prevent diarrheagenic E. coli in the short term, such as improved WASH infrastructure, exclusive breastfeeding, optimal nutrition, and the creation of community awareness.
Cholera
Cholera, caused by the bacterium Vibrio cholerae, is a diarrheal disease that causes devastating epidemics as well as sporadic cases in many settings in SSA that still have challenges with overcrowding, poor WASH infrastructure, and displaced populations. According to the WHO, 1.3–4 million cholera infections occur globally and out of these 21,000−143,000 result in death. In SSA, cholera is still endemic and is responsible for 141,918 infections per year (95% CI 141,538–146,505).52 The bacterium V. cholerae is composed of 206 recognized serogroups,53 two of which (ie serogroups 01 and 0139) cause epidemics. These serogroups are further divided into two distinct serotypes, namely, Classical and El Tor. The V. cholerae Ogawa biotype has been shown to be the most prevalent in the region, while epidemics are mainly caused by Wave 3 atypical O1 El Tor strain, which has been reported to be resistant to multiple antimicrobials,54,55 as shown in Table 3.
 |
Table 3 Antibiotic Resistance Rates of Clinical Vibrio cholerae in SSA |
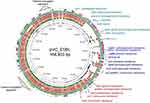
In the past decade, V. cholerae isolates have become resistant to penicillins, aminoglycosides and sulfonamides, quinolones, tetracycline, macrolides, and phenicols,55–59 and more recently several studies have reported strains showing reduced susceptibility to fluoroquinolones and third generation cephalosporins.54,57,58,60 This resistance has been associated with the mutation of housekeeping genes such as gyrA (Ser83Ile) and parC (Ser85Leu),56,60 the presence of mobile genetic elements such as integron class 2,56 and the presence of integrative conjugative elements (ICEs), mainly SXT/R391 (Figure 1). However, clinical and environmental non-O1/O139 V. cholerae have been reported to have low-level resistance to commonly used antimicrobials.60 In Kenya, the extended-spectrum β-lactamase (ESBL)-producing V. cholerae isolates have been found to carry a 168,803-bp putative conjugative plasmid belonging to IncA/C that we designated as pVC_ESBL. The pVC_ESBL plasmid shares extensive sequence identity and synteny with related plasmids (Figure 2), and the incorporation of the class 1 integron and the insertion of the blaCTX-M-15 on the plasmid backbone.
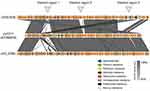 |
Figure 1 Resistance elements inserted into the novel pVC_ESBL plasmid in Kenyan cholera strains. |
Oral rehydration remains the mainstay of management of cholera cases, while antimicrobial use is reserved for the treatment of severe illness. Currently, three WHO pre-qualified oral cholera vaccines (OCVs) – Dukoral®, Shanchol™, and Euvichol® – are administered through mass vaccinations, especially in cholera-endemic areas for the purpose of reducing transmission in endemic settings. In addition to vaccines, improvement of the WASH infrastructure will aid in the control of cholera in SSA.
Typhoid Fever
Typhoid fever is caused by Salmonella enterica serotype Typhi (S. Typhi). Globally, 10.9 million (9.3–12.6 million) cases of typhoid fever occur every year, resulting in nearly 117,000 deaths.65 In SSA, approximately 1.5 million cases are reported annually and 19,900 deaths, accounting for approximately 17% of global deaths.65 In the 1980s in most SSA countries, S. Typhi isolates were fully susceptible to firstline antibiotics, but in the past two decades, multidrug-resistant (defined as resistance to ampicillin, trimethoprim–sulfamethoxazole, and chloramphenicol) S. Typhi has emerged and spread throughout the region.66–68 Because of this resistance, treatment options were revised to include azithromycin, fluoroquinolones, and third-generation cephalosporins. Subsequently, S. Typhi isolates with reduced susceptibility to these antimicrobials have now been reported in Kenya,68,69 South Africa,70 and Congo.71 In a study undertaken in Ibadan, Nigeria, 4.0% of febrile patients with bacteremia were found to be positive for S. Typhi, while 1% had invasive non-typhoidal Salmonella (iNTS) infections.72 All of the Salmonella isolates except one showed multidrugresistance (resistance to at least three antimicrobial classes), with ciprofloxacin non-susceptibility observed in 70.3% and 57% of S.Typhi and iNTS, respectively. Recent data from Kenya show that S. Typhi H58 subgroups circulating in the study setting all had high rates (>70%) of multidrug resistance (MDR) and clearly showed that multidrug-resistant strains predominate and have a high carriage rate in the asymptomatic population.73
Previously, S. Typhi populations were classified into 85 haplotypes (haploid genotypes), but currently, they are categorized into four clusters, 16 clades, and 49 subclades.74 Subclade 4.3.1 (previously haplotype 58) is the predominant MDR genotype in SSA,75 with a prevalence ranging from 60.4% to 97%,73,76–78 which has been associated with not only acute cases but also carriers.73 The resistance in 4.3.1 genotype has been attributed to the presence of translocated regions containing plasmids such as IncHI1 and IncN (subtype PST3),66,69,73,77,78 and the presence of chromosomal mutations in the quinolone resistance-determining regions (QRDRs) of genes parC, gyrA, gyrB, and gyrS.73,78
One of the key factors that has contributed greatly to the AMR of S. Typhi in SSA is the challenges encountered in diagnosis. First, treatment is often prescribed based on symptoms; this is due to the unavailability of microbiology laboratories to perform culture, which is the gold standard but requires invasive sampling, adequate infrastructure, and skilled staff. Often, typhoid symptoms are compounded by or confused with other febrile illnesses such as malaria, owing to their similarity. Secondly, clinicians rely on the Widal test, which is accessible and cheap, and does not require sophisticated technical knowledge to use. However, the Widal test has been proven to be unreliable as it cross-reacts with other Salmonella serotypes and other enteric bacteria. The test is open to widespread abuse and misinterpretation, titers are often guessed using a rapid slide agglutination without serial dilution/tubes, timing of the test is inaccurate, and serum quantities are not measured (there is often no measuring device), among other challenges.79 The introduction of rapid and easy-to-use diagnostic tools for typhoid fever surveillance, potentially including environmental surveillance, and the improvement of existing methods have been recommended80 as some of the best approaches to facilitate the identification of high-risk areas. Such rapid and easy-to-use tools need to be affordable, applicable to the illnesses, easy to perform anywhere, and safe and easy to store in rural areas, among other key desirable factors.
Antibiotic treatment is still the mainstay of typhoid management. The recommended antimicrobials are fluoroquinolones, third-generation cephalosporins, and azithromycin. Vaccination prophylaxis is key in the control and prevention of typhoid, especially in endemic areas. Three typhoid vaccines are currently recommended by the WHO: 1) typhoid conjugate vaccine (TCV), 2) unconjugated polysaccharide vaccine (Vi-PS), and 3) oral live-attenuated Ty21a vaccine. Table 4 shows the antibiotic resistance rates of S. Typhi from clinical samples as previously reported in various countries in SSA. Based on these studies, the AMR rates of S. Typhi vary in different countries and regions.
 |
Table 4 Antibiotic Resistance Rates of Clinical Salmonella enterica Serotype Typhi Isolates in SSA |
Invasive Non-Typhoidal Salmonellosis
iNTS accounts for 534,600 (409,000–705,000) cases and 77,470 (46,360–122,800) deaths annually worldwide; out of these, 421,600 (316,000–574,100) cases (26.2% attributed to HIV infection) and 66,520 deaths occur in SSA.87 The highest incidence is observed among children aged 0–5 years, followed by the 15–19 and 35–49-year-old age groups. The major predisposing factors for iNTS infection in children are malnutrition, malaria parasitemia, and underdeveloped immunity, while in adults, HIV infection is the primary factor.88–91 Other significant host factors are anemia and sickle cell.92,93 In urban areas where iNTS is endemic, malnutrition plays a key role in adult infection, especially where exposure to malaria is minimal.89 In SSA, the incidence of iNTS disease in children ranges from 166 to 568/100,000 cases of person-years of observation (pyo).94 In most parts of SSA, iNTS presents as non-specific febrile illness, which poses a major challenge in diagnosis as it is difficult to distinguish from other febrile illnesses endemic in these settings, such as malaria. In addition, most NTS infections present as invasive disease, especially among children less than 5 years of age and those with underlying immunosuppressive conditions.95 Of all the NTS serotypes, S. Typhimurium and S. Enteritidis contribute the highest proportion of invasive disease. In Kenya, for instance, of all iNTS isolates, 33.6% were S. Typhimurium while 22.8% were S. Enteriditis.96 A review published in 201793 found that since 1966, NTS has been a major cause of bacteremia in 33 out of 54 African countries, accounting for up to 39% of community-acquired bloodstream infections in the region and resulting in an average case fatality rate of 19%. Increasing rates of MDR in iNTS are of great concern, and the situation is even more acute in SSA settings where alternative options for effective treatment are being rapidly eroded. High levels of multidrug-resistant iNTS have been reported in Kenya,97–99 and in other parts of SSA, including DRC,100,101 Ghana,102 and Uganda,103 posing a major challenge to treatment and management options for severe disease.
Invasive strains of multidrug-resistant NTS circulating in SSA are predominantly S. Typhimurium ST313 and S. Enteritidis ST11, although S. Dublin ST10 has only been associated with resistance in West Africa.96,104 These strains harbor the notorious Tn21 transposon on an incF plasmid (pSLT-BT), which contains a plethora of resistance genes. In addition, resistance to third generation cephalosporins has been attributed to the presence of ESBL-encoding genes such as blaCTX-M, while non-susceptibility to fluoroquinolones has been associated with mutation of the gyrA gene.104 Multidrug-resistant S. Typhimurium ST313 has now been reported from several countries in SSA, including Kenya,105 Malawi,67 Central African Republic, Senegal, Burkina Faso,106 DRC,107 Nigeria,108 Ghana,102 South Africa, and Mozambique,79,109 where iNTS is endemic and produces septicemia in the absence of gastroenteritis.
In another study of the genetic characteristics and transmission patterns associated with multidrug-resistant iNTS serovars across SSA,104 multiple multidrug-resistant iNTS serovar sequence types were identified as circulating in several countries that were sampled. High carriage rates observed in endemic sites in Kenya of MDR ST313110 are likely to provide a reservoir for community spread of infection to vulnerable populations, adding new challenges in the control and prevention of iNTS disease.
Efforts to increase access to diagnostic services are required to improve patient outcomes and also to estimate disease burden. There are currently no rapid diagnostic tests available for the detection of iNTS, and the development of such tests would greatly facilitate the study and management of iNTS disease in SSA. The development of novel diagnostic tools is challenging owing to the diversity of NTS serotypes, but the use of locally targeted lipopolysaccharide antigen is likely to be useful.79,111
Third generation cephalosporins (ceftriaxone) and fluoroquinolones (ciprofloxacin) are the mainstay of iNTS treatment. Non-fluoroquinolone-susceptible cases are usually treated with azithromycin. Currently, no vaccine is available for the long-term prevention and control of iNTS, but several vaccine candidates are under development. Sustained disease surveillance of multidrug-resistant iNTS organisms is necessary, and there is a need to speed up iNTS vaccines. Tables 5 and 6 show the antibiotic resistance rates of S. Typhimurium and S. Enteritidis, respectively, as reported in various studies conducted in SSA.
 |
Table 5 Antimicrobial Resistance Rates of Clinical Salmonella enterica Serotype Typhimurium Isolates in SSA |
 |
Table 6 Antimicrobial Resistance Rates of Clinical Salmonella enterica Serotype Enteritidis Isolates in SSA |
Multidrug-Resistant Klebsiella pneumoniae-Associated Infections
Klebsiella pneumoniae is the third leading causative agent for healthcare-associated infections, after E. coli and S. aureus.117,118 It is an opportunistic pathogen which causes infections on otherwise sterile sites, for example septicemia, bacteremia, and infections of the respiratory tract, urinary tract, and central nervous system. Of concern are outbreaks in neonatal units, where the infection rates are 3–20-fold higher than rates in in high-income countries.118 More than 50% of healthcare-associated infections associated with K. pneumoniae are reportedly ESBL producers,119 and hence the WHO has recognized ESBL and carbapenem-resistant K. pneumoniae as public health priorities.120 In SSA, precise burden estimates for resistant K. pneumoniae are lacking and the available data are generated in referral/research/university hospitals, the context of which does not represent the broad range of healthcare settings.118 These data, however, indicate that the rates are increasing.
In SSA, high-risk populations are newborns and patients admitted to intensive care units (ICUs), especially those with underlying comorbidities.121,122 The main driving factors for resistant K. pneumoniae are poor hand hygiene among healthcare workers, contaminated medical equipment, and poor waste management practices in hospitals.123,124 These drivers are associated with inadequate supply of equipment, poor infrastructure relating to infection control, and understaffed healthcare settings, which result in overcrowded wards and high patient load.
Based on the seven-gene multilocus sequence typing, K. pneumoniae is classified into various sequence types (STs). ST15, ST17, and ST70 have been associated with ESBL strains, and have been reported in Eastern Africa and Southern Africa.125–127 ST152, ST340 (Clonal Group 258), and ST307 have been associated with carbapenem resistance and have only been reported in Southern Africa.128–130 These resistant strains have been shown to harbor multiple resistance genes encoded on the chromosome or on plasmids; namely, bla SHV, bla LEN, bla OKP, bla CTX-M, bla OXA, bla TEM, bla KPC, bla NDM, bla KPC, and bla VIM.128–130
To control and prevent hospital-associated resistant K. pneumoniae, effective infection control measures will have to be undertaken to ensure hand hygiene, proper use of personal protective equipment, injection safety, and proper disinfection of hospital surfaces and sterilization of medical equipment. Education and training of healthcare workers is crucial, with an aim to create awareness on effective infection control and its impact on nosocomial infections, and antibiotic stewardship. In addition, the implementation of local and national surveillance systems is required in the region to give precise burden estimates.
Carbapenem-Resistant Acinetobacter baumannii
Acinetobacter species are Gram-negative bacteria commonly pervasive in the environment (soil and water).131,132 Of all the 60 known Acinetobacter species, Acinetobacter baumannii complex, which consists of Acinetobacter pittii, Acinetobacter nosocomialis, and Acinetobacter baumannii (A. baumannii), is the most clinically relevant.132,133 Acinetobacter baumannii accounts for the majority (approximately 90%) of clinical infections, followed by A. pittii and A. nosocomialis.134 Acinetobacter baumannii is an opportunistic bacterial pathogen mainly associated with hospital-acquired infection;135 however, community-acquired infections are increasing.136 It is a Gram-negative, catalase-positive, oxidase-negative, non-fermenting coccobacillus that is aerobic, pleomorphic, and non-motile. It has a high incidence among immunocompromised individuals, particularly those who have experienced a prolonged (>90-day) hospital stay.135,136 Other risks that exacerbate these infections include prolonged exposure to antibiotics, mechanical ventilation, and central venous catheter use.131 Acinetobacter baumannii is often implicated in bloodstream, urinary tract, pneumonia, skin and soft tissue infections, and meningitis.138,139
Owing to its AMR,140 A. baumannii is one of the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) that pose a huge global health challenge because of their MDR.139 Carbapenem-resistant Acinetobacter baumannii (CRAB) is currently ranked first on the WHO list of critical pathogens urgently in need of new antibiotic development.141 The global prevalence of A. baumannii, its widespread antibiotic resistance, and most recently, the unforeseen emergence and spread of clinical isolates resistant to colistin, the last resort in current antimicrobial therapy, all necessitate its inclusion in national surveillance systems and national action plans.141,142 According to the WHO-GLASS report 2021,34 A. baumannii and E. coli were the most frequently reported pathogens globally in the AMR surveillance systems. A review published in 2021 that looked into data on A. baumannii from the 29 countries in the low-income country (LIC) group showed that there are very few reports coming out of this group of countries.141 Out of the 29 countries, only 16 countries reported the detection of A. baumannii within their territories at least once in the last 21 years. Accordingly, 11 out of the 29 have no A. baumannii reports at all in the three searched databases. The country with the highest number of reports was Ethiopia, with 10 reports. Interestingly, high rates of carbapenem resistance have been reported in other studies of comparable countries in terms of economic development (reaching 100% in some instances).141 The modest numbers of A. baumannii reported in LICs could therefore just be a reflection of the low numbers of isolates investigated in many of these studies. Accordingly, it could be very difficult to obtain a good estimate of the prevalence of carbapenem resistance detected in the LICs.141
Carbapenems have been the preferred antibiotics for the treatment and management of multidrug-resistant A. baumannii; however, previous use has led to the emergence of and increase in carbapenem-resistant A. baumannii.143 Acinetobacter baumannii has a high propensity for intrinsic resistance to different antibiotics and the ability to acquire different antibiotic resistance genes. The choice of carbapenem as a marker for MDR is based on its correlation with broad-range coresistance to different antibiotics.144 Carbapenem resistance in A. baumannii is predominantly mediated by carbapenem-hydrolyzing Ambler class D β-lactamases (CHDLs), especially the oxacillinase (OXA) type, and Ambler class B metallo-β-lactamases (MBLs).145 Studies in SSA have detected CHDL genes, including blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, blaOXA-143-like, and blaOXA-235-like, among others, and MBL genes (blaIMP, blaVIM, and blaNDM) in carbapenem-resistant A. baumannii.145–150
In 2019, A. baumannii infections were the fifth most common cause of deaths attributable to antimicrobial-resistant bacteria globally, with 57,700 mortalities attributable to carbapenem-resistant A. baumannii.151 The global percentage of resistance to carbapenems of A. baumannii varies geographically in different continents, regions, and countries.152 Several recent studies in SSA have reported varying levels of carbapenem resistance in A. baumannii (Table 7). A 2018 study reported that in East Africa, A. baumannii had the highest prevalence (23%) of carbapenem resistance compared to other multidrug-resistant bacteria.148
 |
Table 7 Antimicrobial Resistance Rates of Clinical Acinetobacter baumannii in SSA |
There are currently no vaccines available against A. baumannii; however, several studies have investigated the efficacy of different vaccine candidates through preclinical trials. This highlights the need for vaccine research as one of the measures for managing A. baumannii infections.152
Staphylococcus aureus
Staphylococcus aureus infections have long been a public health concern, and particularly those associated with methicillin-resistant Staphylococcus aureus (MRSA), which continues to threaten modern healthcare. An MRSA study conducted in the European Union, published in 2019, estimates an annual number of disability-adjusted life-years (DALYs) at 148,727 (131,757–166,361) per 1,000,000 population, infections at 112,782 (103,186–122,006), and attributable deaths at 6810 (6096–7559).156 While surveillance systems are in place in developed countries, they are lacking in developing countries, including those in the SSA region, and most data are from single-center studies. Nevertheless, community and healthcare-associated MRSA has been reported in most countries, including Kenya, Uganda, Nigeria, South Africa, Ghana, Mozambique, Ethiopia, and Tanzania, where the MRSA prevalence ranges from 1.25% to 53.4%.157–164 The most common risk factors associated with MRSA infection are HIV infection, prior antibiotic use, ICU admission, prior or prolonged hospitalization, indwelling catheters, invasive procedures, open wounds and burns, residency at long-term care facilities, mechanical ventilation, renal failure, and MRSA colonization history.165,166 Laboratory diagnosis is by culture, serology, and molecular assays. Often, laboratory equipment is available in tertiary facilities, but not in primary healthcare facilities.
The choice of treatment for S. aureus infections is dependent upon susceptibility, site, and origin of infection, ie community or hospital-acquired. Various antimicrobials are recommended, for either monotherapy or combined therapy, including methicillin, oxacillin, erythromycin, tetracycline, clindamycin, rifampicin, vancomycin, linezolid, teicoplanin, trimethoprim/sulfamethoxazole, daptomycin, quinupristin/dalfopristin, amikacin, ciprofloxacin, chloramphenicol, mupirocin phosphomycin, fusidic acid, and tigecycline. Resistance to these antimicrobials has emerged (Table 8), and the most common resistance phenotype among clinical S. aureus is erythromycin, tetracycline, and co-trimoxazole. This has been attributed to the presence of several genes; aac(6’)-Ie-aph(2”)-Ia, aph(3’)-IIIa, ant(4’)-Ia, apH3, and aacA-aphD, conferring resistance to aminoglycosides; tetK/M genes against tetracycline; dfrG/dfrA genes against trimethoprim; ermA, ermC, or msrA genes against clindamycin; and gyrA and grlA genes against fluoroquinolones.158,167,168 A majority of MRSA isolates have been shown to innately harbor mecA and blaZ genes, which encode for a range of penicillinases. Moreover, the most prevalent genotypes have been demonstrated to be ST5, ST8, ST15, ST88, ST152, ST241, and ST239.162,169–171
 |
Table 8 Antimicrobial Resistance Rates of Staphylococcus aureus in SSA |
With the rise of AMR, research and development for vaccines has grown considerably; however, most vaccine candidates have failed to pass phase III clinical trials and, therefore, no vaccine is currently licensed. In the short term, proper implementation of infection control and prevention measures will be key in reducing and tackling S. aureus infections, and especially those caused by MRSA.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a Gram-negative bacterium commonly found in water and soil.173 It is an opportunistic pathogen commonly associated with hospital-acquired infections, particularly bloodstream infections, urinary tract infections, and pneumonia, resulting in considerable morbidity and mortality. The risk factors often associated with P. aeruginosa infections include prior hospitalization, ICU stay, and previous use of antibiotics.174
Pseudomonas aeruginosa has an intrinsic ability to develop resistance to frequently used antibiotics, which often leads to treatment failure. Of concern is its resistance to beta-lactams,175 especially carbapenems, as they are often used for empiric treatment of last resort.176 The WHO has listed carbapenem-resistant P. aeruginosa as one of the three critical global priority pathogens that require development of new antibiotics.144 It is estimated that in 2019, antimicrobial-resistant P. aeruginosa accounted for 84,600 mortalities, with carbapenem-resistant P. aeruginosa accounting for 38,100 deaths globally.151 The prevalence of carbapenem-resistant P. aeruginosa ranges from 6% to 34.8%, as reported in various studies conducted in Uganda, Ghana, South Africa, Zambia, Nigeria, and Ethiopia.176–181
Although P. aeruginosa has an intrinsic resistance to several antibiotics, it can develop resistance through the acquisition of resistance genes.182,183 Various carbapenemase-encoding genes have been observed: blaIMP, blaSPM, blaVIM, blaNDM, blaOXA-23, blaOXA-48 blaSIM, blaAIM, blaBIC-1, blaNMC-A, and blaOXA-58. The co-occurrence of class I and class II integrons has also been reported, which possess a variety of resistance genes, such as ant(4′)-IIb, dfrA, aadAB, qnrVC1, gyrA, and parC.147,177,179,181,182,184–187
One Health Approach to Tackle AMR
The One Health approach is an important concept that recognizes the inseparable links between human, animal, and environmental health.188 Although AMR surveillance studies have mainly focused on humans and animals, the environment is a critical aspect in the control of AMR. Residues of antibiotics used in humans and animals often end up in the environment,189,190 especially water sources, leading to the persistence of antibiotic residues and resistance genes in the environment. Consequently, hospital waste, farm, industrial, human waste, and waste water treatment plants become reservoirs and dissemination pathways of AMR.189 Therefore, as part of the control measures applied to combat AMR, it would be imperative to identify the environmental reservoirs and dissemination pathways of AMR, and undertake continuous AMR surveillance in the environment191 and improvement of WASH infrastructure. The importance of the One Health approach is clearly demonstrated in the global action plan (GAP) on AMR.
Implementation of the Global Action Plan and National Action Plans in Sub-Saharan Africa
The GAP to combat AMR was adopted in 2015 by 194 member states of the WHO through decisions made at the 68th session of the World Health Assembly, by the Food and Agriculture Organization (FAO) Governing Conference, and by delegates at the World Assembly of World Organisation for Animal Health, formerly the Office International des Epizooties (OIE). This followed reports of alarming rates of resistance to healthcare and community-acquired infections as well as reports of resistance in agriculture, livestock, and fisheries. Considering the transboundary challenges of AMR, world leaders attending the United Nations General Assembly in 2016 recognized the need for collaboration in tackling AMR.27 Based on the framework of the GAP, countries were urged to develop national action plans (NAPs) to tackle AMR by 2017.192 The aim of the GAP is to ensure the successful treatment and prevention of infectious diseases with effective, safe medicines that are quality assured, used in a responsible way, and accessible to all who need them.5 The GAP lays out important activities based on five objectives to be undertaken through a continuous multisectoral approach, aimed at minimizing the impact of AMR globally.
The five objectives are: 1) to improve awareness and understanding of AMR; 2) to strengthen knowledge through surveillance and research; 3) to reduce the incidence of infection; 4) to optimize the use of antimicrobial agents; and 5) to ensure sustainable investment in countering AMR.5,27 To underscore the importance of the One Health (human health, animal health, and agriculture) approach in combating AMR, a tripartite collaboration that involves the WHO, FAO, and OIE was initiated. The role of the tripartite collaboration is to encourage progress in the development, costing, and harmonization of NAPs, while monitoring and evaluating the progress made by the member states.
Globally, two-thirds of the 194 WHO member states are reported to have developed NAPs.192 The overall performance of SSA countries in the implementation of NAPs has been reported to be inadequate. Between 2016 and 2017, only 14 countries in the WHO Africa region reported having developed NAPs, while nine could monitor antimicrobial consumption and use.27 A more recent study found that 11 (25%) out of 44 SSA countries had NAPs on AMR, with 32% and 2% performing routine AMR surveillance on clinical and veterinary pathogens, respectively.193 Fifty-nine percent of the countries reported having a dedicated laboratory that serves as the National Reference Laboratory for AMR. By February 2022, a library of existing, publicly available NAPs on AMR compiled by the WHO (https://www.who.int/teams/surveillance-prevention-control-AMR/national-action-plan-monitoring-evaluation/library-of-national-action-plans) included 15 SSA countries with fully developed NAPs: Burkina Faso, Eritrea, Eswatini, Ethiopia, Ghana, Kenya, Liberia, Malawi, Mauritius, Nigeria, Sierra Leone, South Africa, Tanzania, Zambia, and Zimbabwe.
The Eastern SSA region has been reported to have the highest number of countries implementing NAPs, followed by Southern, Western, and Central SSA. A high number of East African countries was found to be undertaking AMR surveillance in human and animal pathogens.193,194 Southern African countries were found to have the highest rate of developing antimicrobial stewardship guidelines and antimicrobial legislation. Although most SSA countries have developed or are in the process of developing NAPs on AMR, Guinea and Angola were reported to be missing NAPs on AMR.
Some of the countries with the best implementation of NAPs in SSA are Tanzania, Ethiopia, Kenya, South Africa, Zambia, and Ghana. In Kenya, for example, owing to the multisectoral nature of AMR and the need for a concerted effort across sectors, the NAP was developed through a multisectoral One Health approach and endorsed jointly by the cabinet secretaries responsible for human health, animal health, and crop production. However, elsewhere, Mozambique was found to have inadequate implementation of NAPs, especially in collaboration with different sectors.194 According to the findings of a study by Iwu and Patrick, the performance of SSA countries in the different thematic areas of the GAP was 38±12% in awareness creation and training, 18±13% in surveillance, 33±13% in prevention and control infections, 28±23% in optimal use of antimicrobial agents, and 59±12% in multisector and One Health collaboration.194 In several SSA countries, including Kenya, the Fleming Fund195 has greatly facilitated the implementation of the NAPs, especially raising awareness, ensuring an expanded antimicrobial stewardship framework to optimize antimicrobial use, increase AMR and antimicrobial use surveillance, monitoring, and data sharing, and the implementation of infection control and prevention strategies.
Challenges in Implementation of NAPs in Sub-Saharan Africa
Although some progress has been in the implementation of NAPs in SSA, several challenges still hinder effective implementation. Whilst most of the SSA countries have developed NAPs, their full implementation is the greatest challenge. Some of the challenges that hinder the implementation of NAPs are: 1) a lack of political commitment, 2) inadequate funding, 3) poor coordination and collaboration between the different sectors, 4) inadequate surveillance data, 5) poor infrastructure and health systems, 6) staffing shortages, and 7) a lack of monitoring and quality assurance systems.196,197
Political commitment is a key factor in the successful implementation of NAPs on AMR.198 Consequently, the absence of political will often translates into poor or lacking resource allocation for the implementation of NAPs, and inadequate regulatory policies and enforcement. The lack of commitment by policymakers or political leaders could be attributed to the lack of awareness and appreciation of the impact of AMR on the quality of human life. Inadequate funding/resource allocation in SSA is attributable to limited resources, and a lack of awareness and/or commitment by the private and public sectors. The implementation of NAPs is a resource-intensive activity; therefore, a lack of continuous funding results in a paucity of surveillance data, staffing inadequacy, and poor infection prevention and control. The scarcity of financial resources in most SSA countries means that funding of NAPs is not prioritized, since government spending on health is low. In 2001, African governments through the Abuja declaration committed to spending 15% of government budgets on public health. However, between 2002 and 2014, the percentage of government spending on health decreased in approximately half of SSA countries.199 It is important to note that a WHO report estimated that between 20% and 40% of health budgets are wasted through inefficiencies and corruption.200
Effective implementation of NAPs requires multisectoral collaboration and coordination between human health, animal health, and environmental/food sectors. The multisectoral collaboration ought to involve governments, healthcare workers, researchers, academicians, veterinarians, farmers, and the private sector.194 However, owing to the absence of coordinating agencies, coordination among the different sectors is problematic. One of the challenges identified in the implementation of NAPs in Mozambique was the poor collaboration between the different sectors.194
Data on disease epidemiology, AMR, antibiotic consumption, environmental exposure pathways, access to healthcare, morbidity, hospital readmission rates, length of stay in hospital, and mortality rates are critical in informing policy and actions to be undertaken at the country level. However, because of the huge costs associated with research and other data collection, there is a scarcity of data in SSA. Consequently, the absence of these data creates a challenge in the successful implementation of NAPs, since it is difficult to assess whether progress is being made.
Poor WASH infrastructure and healthcare systems are also a challenge in the implementation of NAPs in SSA since they contribute to the spread of antimicrobial-resistant pathogens and poor diagnosis. Proper diagnostic or microbiological laboratory services are critical in ensuring the proper use of antimicrobials; however, poor diagnostic services remain a huge problem in SSA.
Inadequate competent human resources have also been identified as a major challenge in healthcare provision in Africa.194,197,201 Lack of personnel has also been identified as a barrier to antimicrobial stewardship programs.202 The inadequacy of staff working on AMR limits the implementation of NAPs at the country level.
Improvement of Implementation of NAPs Towards Control and Prevention of AMR
To actualize the objectives of the GAPs, there needs to be an improvement in the implementation of NAPs at the country level. While several SSA countries have developed NAPs, these plans have not yet been fully implemented. In an implementation handbook published in 2022,203 the WHO has outlined six steps that would ensure the sustainable implementation of NAPs on AMR: 1) strengthening governance, 2) prioritizing activities for implementation, 3) costing the NAPs’ operational plan, 4) resource mobilization, 5) implementing prioritized activities, and 6) monitoring and evaluation.
To ensure the effective implementation of NAPs, SSA countries need to prioritize the funding of these plans. There is a need to factor in the funding of these plans as part of the health sector. Governments also need to engage the private sector in the mobilization of resources. It is also imperative to have a coordination team that is comprised of expert groups and members from different sectors. The coordination team would be critical in identifying areas of priority at the national level, harmonizing the activities of different NAPs, mobilizing resources, and engaging policymakers. The coordination team would also be imperative for SSA countries in monitoring antimicrobial use and in the continuous surveillance of AMR prevalence and patterns. However, to achieve this, SSA countries need to ensure that there is a capacitated human resource dedicated to the implementation of NAPs. Scientists and academicians also need to undertake research that focuses not only on resistance levels but also on the impact of AMR on the socio-economic aspects of human life. Collaboration among SSA countries would be critical, especially in the areas of coordinated training, legal frameworks, and data-sharing platforms.
Conclusion
AMR continues to be a major challenge in the effective control and management of infectious pathogens. This review has highlighted the AMR rates in different WHO-GLASS and locally important enteric pathogenic bacteria, and the successes and challenges in the implementation of NAPs in sub-Saharan Africa. Countries including Tanzania, Ghana, and South Africa were found to have better implementation of NAPs as a result of the multisectoral collaboration in those countries. Based on the available data, it is important to note that death rates attributable to AMR are highest in SSA, yet the paucity of data in SSA on AMR prevalence and antimicrobial use is a huge impediment to the effective control and management of infectious diseases. The few available data indicate an increase in reports of AMR rates and resistance genes, which could be resulting in treatment failure and increasing mortalities. Therefore, there is a need for SSA countries to ensure continuous data generation, which will be key in informing strategies to be implemented at the country level as different countries have varying resistance levels. It is also imperative that the One Health approach is implemented in the surveillance and control of AMR. In addition to surveillance, effective implementation of infection, prevention and control, hand hygiene, and antimicrobial stewardship programs will be key cost-effective strategies in tackling AMR.
Funding
SK is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [award number R01AI099525] and the Wellcome Trust [grant number 215675-Z-19-Z].
Disclosure
The authors report no conflicts of interest in relation to this work.
References
1. Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):1–8. doi:10.1186/s13756-017-0208-x
2. Morel CM, Alm RA, Årdal C, et al. A one health framework to estimate the cost of antimicrobial resistance. Antimicrob Resist Infect Control. 2020;9(1):1–14. doi:10.1186/s13756-020-00822-6
3. Dadgostar P. Antimicrobial resistance: implications and costs. Infect Drug Resist. 2019;12:3903–3910. doi:10.2147/IDR.S234610
4. Varma JK, Oppong-Otoo J, Ondoa P, et al. Africa Centres for Disease Control and Prevention’s framework for antimicrobial resistance control in Africa. Afr J Lab Med. 2018;7(2):830. doi:10.4102/ajlm.v7i2.830
5. Mendelson M, Matsoso MP. The World Health Organization Global Action Plan for antimicrobial resistance. S Afr Med J. 2015;105(5):325. doi:10.7196/SAMJ.9644
6. Hayat K, Rosenthal M, Gillani AH, et al. Perspective of Pakistani physicians towards hospital antimicrobial stewardship programs: a multisite exploratory qualitative study. Int J Environ Res Public Health. 2019;16(9):1565. doi:10.3390/ijerph16091565
7. Friedman ND, Temkin E, Carmeli Y. The negative impact of antibiotic resistance. Clin Microbiol Infect. 2016;22(5):416–422. doi:10.1016/j.cmi.2015.12.002
8. Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–655. doi:10.1016/S0140-6736(21)02724-0
9. Macintyre A, Wilson-Jones M, Velleman Y. Prevention first: tackling AMR through water, sanitation and hygiene. 2017.
10. Musoke D, Namata C, Lubega GB, et al. The role of environmental health in preventing antimicrobial resistance in low- and middle-income countries. Environ Health Prev Med. 2021;26(1):100. doi:10.1186/s12199-021-01023-2
11. Li G, Bielicki JA, Ahmed ANU, et al. Towards understanding global patterns of antimicrobial use and resistance in neonatal sepsis: insights from the NeoAMR network. Arch Dis Child. 2020;105(1):26–31. doi:10.1136/archdischild-2019-316816
12. Fadare JO, Ogunleye O, Iliyasu G, et al. Status of antimicrobial stewardship programmes in Nigerian tertiary healthcare facilities: findings and implications. J Glob Antimicrob Resist. 2019;17:132–136. doi:10.1016/j.jgar.2018.11.025
13. Parry CM, Wijedoru L, Arjyal A, et al. The utility of diagnostic tests for enteric fever in endemic locations. Expert Rev Anti Infect Ther. 2011;9(6):711–725. doi:10.1586/eri.11.47
14. O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. 2016.
15. Ferri M, Ranucci E, Romagnoli P, et al. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–2876. doi:10.1080/10408398.2015.1077192
16. Taylor J, Hafner M, Yerushalmi E, et al. Estimating the Economic Costs of Antimicrobial Resistance. Cambridge, UK: Model and Results (RAND Corporation); 2014.
17. McEwen SA, Collignon PJ, Aarestrup FM, Schwarz S, Shen J, Cavaco L. Antimicrobial resistance: a one health perspective. Microbiol Spectr. 2018;6(2). doi:10.1128/microbiolspec.ARBA-0009-2017
18. Sartelli M, Catena F, Catena F, et al. Antibiotic use in low and middle-income countries and the challenges of antimicrobial resistance in surgery. Antibiotics. 2020;9(8):497. doi:10.3390/antibiotics9080497
19. Van TTH, Yidana Z, Smooker PM, et al. Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J Glob Antimicrob Resist. 2020;20:170–177. doi:10.1016/j.jgar.2019.07.031
20. Chokshi A, Sifri Z, Cennimo D, et al. Global contributors to antibiotic resistance. J Glob Infect Dis. 2019;11(1):36. doi:10.4103/jgid.jgid_110_18
21. Spellberg B. The maturing antibiotic mantra:“shorter is still better”. J Hosp Med. 2018;13(5):
22. Ateshim Y, Bereket B, Major F, et al. Prevalence of self-medication with antibiotics and associated factors in the community of Asmara, Eritrea: a descriptive cross sectional survey. BMC Public Health. 2019;19(1):1–7. doi:10.1186/s12889-019-7020-x
23. Yeika EV, Ingelbeen B, Kemah BL, et al. Comparative assessment of the prevalence, practices and factors associated with self‐medication with antibiotics in Africa. Trop Med Int Health. 2021;26(8):862–881. doi:10.1111/tmi.13600
24. Rather IA, Kim B-C, Bajpai VK, et al. Self-medication and antibiotic resistance: crisis, current challenges, and prevention. Saudi J Biol Sci. 2017;24(4):808–812. doi:10.1016/j.sjbs.2017.01.004
25. Simon B, Kazaura M. Prevalence and factors associated with parents self-medicating under-fives with antibiotics in Bagamoyo District Council, Tanzania: a cross-sectional study. Patient Prefer Adherence. 2020;14:1445–1453. doi:10.2147/PPA.S263517
26. Browne AJ, Chipeta MG, Haines-Woodhouse G, et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planetary Health. 2021;5(12):e893–e904. doi:10.1016/S2542-5196(21)00280-1
27. WHO. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation. WHO; 2018.
28. Van Boeckel TP, Pires J, Silvester R, et al. Global trends in antimicrobial resistance in animals in low-and middle-income countries. Science. 2019;365(6459):eaaw1944. doi:10.1126/science.aaw1944
29. Manyi-Loh C, Mamphweli S, Meyer E, et al. Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 2018;23(4):795. doi:10.3390/molecules23040795
30. de Mesquita Souza Saraiva M, Lim K, Do Monte DFM, et al. Antimicrobial resistance in the globalized food chain: a One Health perspective applied to the poultry industry. Braz J Microbiol. 2022;53(1):465–486. doi:10.1007/s42770-021-00635-8
31. Mshana SE, Sindato C, Matee MI, et al. Antimicrobial use and resistance in agriculture and food production systems in Africa: a systematic review. Antibiotics. 2021;10(8):976. doi:10.3390/antibiotics10080976
32. Collignon P, Beggs JJ, Walsh TR, et al. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planetary Health. 2018;2(9):e398–e405. doi:10.1016/S2542-5196(18)30186-4
33. World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020. Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization; 2020.
34. World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021. Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization; 2021.
35. Frost I, Van Boeckel TP, Pires J, et al. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med. 2019;26(8):taz036. doi:10.1093/jtm/taz036
36. Troeger C, Blacker BF, Khalil IA, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1211–1228. doi:10.1016/S1473-3099(18)30362-1
37. Saka HK, Dabo NT, Muhammad B, et al. Diarrheagenic Escherichia coli pathotypes from children younger than 5 years in Kano State, Nigeria. Front Public Health. 2019;7:348. doi:10.3389/fpubh.2019.00348
38. Mabika R, Liabagui S, Kenguele H, et al. Molecular prevalence and epidemiological characteristics of diarrheagenic E. coli in children under 5 years old in the City of Koula-Moutou, East-Central Gabon. Open J Med Microbiol. 2021;11:157–175. doi:10.4236/ojmm.2021.113013
39. Somda NS, Bonkoungou I, Cheikna Z, et al. Prevalence of Escherichia coli virulence genes in patients with diarrhoea in Ouagadougou, Burkina Faso. African J Clin Exp Microbiol. 2017;18:179. doi:10.4314/ajcem.v18i4.1
40. Webale MK, Wanjala C, Guyah B, et al. Epidemiological patterns and antimicrobial resistance of bacterial diarrhea among children in Nairobi City, Kenya. Gastroenterol Hepatol Bed Bench. 2020;13(3):238–246.
41. Msolo L, Iweriebor BC, Okoh AI. Antimicrobial resistance profiles of diarrheagenic E. coli (DEC) and Salmonella species recovered from diarrheal patients in selected rural communities of the Amathole District Municipality, Eastern Cape Province, South Africa. Infect Drug Resist. 2020;13:4615–4626. doi:10.2147/IDR.S269219
42. Seidman JC, Johnson LB, Levens J, et al. Longitudinal comparison of antibiotic resistance in diarrheagenic and non-pathogenic Escherichia coli from young Tanzanian children. Front Microbiol. 2016;7:1420. doi:10.3389/fmicb.2016.01420
43. Mahazu S, Prah I, Ayibieke A, et al. Possible dissemination of Escherichia coli sequence type 410 closely related to B4/H24RxC in Ghana. Front Microbiol. 2021;12:770130. doi:10.3389/fmicb.2021.770130
44. Omolajaiye SA, Afolabi KO, Iweriebor BC, Quinn FD. Pathotyping and antibiotic resistance profiling of Escherichia coli isolates from children with acute diarrhea in Amatole District Municipality of Eastern Cape, South Africa. Biomed Res Int. 2020;2020:4250165. doi:10.1155/2020/4250165
45. Ayibieke A, Sato W, Mahazu S, et al. Molecular characterisation of the NDM-1-encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS One. 2018;13(12):e0209623. doi:10.1371/journal.pone.0209623
46. Ouédraogo AS, Compain F, Sanou M, et al. First description of IncX3 plasmids carrying blaOXA-181 in Escherichia coli clinical isolates in Burkina Faso. Antimicrob Agents Chemother. 2016;60(5):3240–3242. doi:10.1128/AAC.00147-16
47. Prah I, Ayibieke A, Mahazu S, et al. Emergence of oxacillinase-181 carbapenemase-producing diarrheagenic Escherichia coli in Ghana. Emerg Microbes Infect. 2021;10(1):865–873. doi:10.1080/22221751.2021.1920342
48. Igwe JC, Onaolapo JA, Kachallah M, et al.. Molecular characterization of extended spectrum β-lactamase genes in clinical E. coli isolates. J Biomed Eng. 2014;2014. Doi:10.4236/jbise.2014.75030
49. Isendahl J, Turlej-Rogacka A, Manjuba C, et al. Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS One. 2012;7(12):e51981. doi:10.1371/journal.pone.0051981
50. Negeri AA, Seyoum ET, Ibrahim RA, et al. Antimicrobial resistance profile of Escherichia coli isolates recovered from diarrheic patients at Selam Health Center, Addis Ababa, Ethiopia. Afr J Microbiol Res. 2019;13(26):457–463. doi:10.5897/AJMR2018.9030
51. World Health Organization. WHO Preferred Product Characteristics for Vaccines Against Enterotoxigenic Escherichia Coli. Geneva: World Health Organization; 2021.
52. Lessler J, Moore SM, Luquero FJ, et al. Mapping the burden of cholera in sub-Saharan Africa and implications for control: an analysis of data across geographical scales. Lancet. 2018;391(10133):1908–1915. doi:10.1016/S0140-6736(17)33050-7
53. Li Z, Lu X, Wang D, et al. Genomic comparison of serogroups O159 and O170 with other Vibrio cholerae serogroups. BMC Genom. 2019;20(1):1–13. doi:10.1186/s12864-018-5379-1
54. Smith AM, Njanpop-Lafourcade B-M, Mengel MA, et al. Comparative characterization of Vibrio cholerae O1 from five sub-Saharan African countries using various phenotypic and genotypic techniques. PLoS One. 2015;10(11):e0142989. doi:10.1371/journal.pone.0142989
55. Adewale AK, Pazhani GP, Abiodun IB, et al. Unique clones of Vibrio cholerae O1 El Tor with Haitian type ctxB allele implicated in the recent cholera epidemics from Nigeria, Africa. PLoS One. 2016;11(8):e0159794. doi:10.1371/journal.pone.0159794
56. Thompson CC, Freitas FS, Marin MA, et al. Vibrio cholerae O1 lineages driving cholera outbreaks during seventh cholera pandemic in Ghana. Infect Genet Evol. 2011;11(8):1951–1956. doi:10.1016/j.meegid.2011.08.020
57. Danso EK, Asare P, Otchere ID, et al. A molecular and epidemiological study of Vibrio cholerae isolates from cholera outbreaks in southern Ghana. PLoS One. 2020;15(7):e0236016. doi:10.1371/journal.pone.0236016
58. Feglo PK, Sewurah M. Characterization of highly virulent multidrug resistant Vibrio cholerae isolated from a large cholera outbreak in Ghana. BMC Res Notes. 2018;11(1):1–6. doi:10.1186/s13104-017-2923-z
59. Sambe-Ba B, Diallo MH, Seck A, et al.. Identification of atypical El TorV. cholerae O1 ogawa hosting SXT element in Senegal, Africa. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.00748
60. Marin MA, Thompson CC, Freitas FS, et al. Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS Negl Trop Dis. 2013;7(2):e2049. doi:10.1371/journal.pntd.0002049
61. Iramiot JS, Rwego IB, Kansiime C, et al. Epidemiology and antibiotic susceptibility of Vibrio cholerae associated with the 2017 outbreak in Kasese district, Uganda. BMC Public Health. 2019;19(1):1–9. doi:10.1186/s12889-019-7798-6
62. Garbati M, Baba A, Mursal A, et al. Circulating serotypes and antimicrobial susceptibility pattern of Vibrio cholerae isolates from insurgency-stricken Maiduguri, northeastern Nigeria. Sub Saharan Afr J Med. 2019;6(1):21–26. doi:10.4103/ssajm.ssajm_12_19
63. Awuor SO, Omwenga EO, Daud II. Geographical distribution and antibiotics susceptibility patterns of toxigenic Vibrio cholerae isolates from Kisumu County, Kenya. Afr J Prim Health Care Fam Med. 2020;12(1):e1–e6. doi:10.4102/phcfm.v12i1.2264
64. Dengo-Baloi LC, Semá-Baltazar CA, Manhique LV, et al. Antibiotics resistance in El Tor Vibrio cholerae 01 isolated during cholera outbreaks in Mozambique from 2012 to 2015. PLoS One. 2017;12(8):e0181496. doi:10.1371/journal.pone.0181496
65. Stanaway JD, Reiner RC, Blacker BF, et al. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(4):369–381. doi:10.1016/S1473-3099(18)30685-6
66. Baltazar M, Ngandjio A, Holt KE, et al. Multidrug-resistant Salmonella enterica serotype typhi, Gulf of Guinea region, Africa. Emerg Infect Dis. 2015;21(4):655. doi:10.3201/eid2104.141355
67. Feasey NA, Gaskell K, Wong V, et al. Rapid emergence of multidrug resistant, H58-lineage Salmonella typhi in Blantyre, Malawi. PLoS Negl Trop Dis. 2015;9(4):e0003748. doi:10.1371/journal.pntd.0003748
68. Park SE, Pham DT, Boinett C, et al. The phylogeography and incidence of multi-drug resistant typhoid fever in sub-Saharan Africa. Nat Commun. 2018;9(1):5094. doi:10.1038/s41467-018-07370-z
69. Mutai WC, Waiyaki PG, Kariuki S, et al. Plasmid profiling and incompatibility grouping of multidrug resistant Salmonella enterica serovar Typhi isolates in Nairobi, Kenya. BMC Res Notes. 2019;12(1):1–6. doi:10.1186/s13104-019-4468-9
70. Keddy KH, Smith AM, Sooka A, et al. Fluoroquinolone-resistant typhoid, South Africa. Emerg Infect Dis. 2010;16(5):879. doi:10.3201/eid1605.091917
71. Lunguya O, Lejon V, Phoba M-F, et al. Salmonella typhi in the Democratic Republic of the Congo: fluoroquinolone decreased susceptibility on the rise. PLoS Negl Trop Dis. 2012;6(11):e1921. doi:10.1371/journal.pntd.0001921
72. Popoola O, Kehinde A, Ogunleye V, et al. Bacteremia among febrile patients attending selected healthcare facilities in Ibadan, Nigeria. Clin Infect Dis. 2019;69(Suppl 6):S466–s73. doi:10.1093/cid/ciz516
73. Kariuki S, Dyson ZA, Mbae C, et al. Multiple introductions of multidrug-resistant typhoid associated with acute infection and asymptomatic carriage, Kenya. Elife. 2021;10:e67852. doi:10.7554/eLife.67852
74. Wong VK, Baker S, Connor TR, et al. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827. doi:10.1038/ncomms12827
75. Wong VK, Baker S, Pickard DJ, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47(6):632–639. doi:10.1038/ng.3281
76. Kariuki S, Revathi G, Kiiru J, et al. Typhoid in Kenya is associated with a dominant multidrug-resistant Salmonella enterica serovar Typhi haplotype that is also widespread in Southeast Asia. J Clin Microbiol. 2010;48(6):2171–2176. doi:10.1128/JCM.01983-09
77. Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, et al. Genomic signature of multidrug-resistant Salmonella enterica serovar Typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol. 2015;53(1):262–272. doi:10.1128/JCM.02026-14
78. Mashe T, Leekitcharoenphon P, Mtapuri-Zinyowera S, et al. Salmonella enterica serovar Typhi H58 clone has been endemic in Zimbabwe from 2012 to 2019. J Antimicrob Chemother. 2021;76(5):1160–1167. doi:10.1093/jac/dkaa519
79. Crump JA, Heyderman RS. A perspective on invasive Salmonella disease in Africa. Clin Infect Dis. 2015;61(suppl_4):S235–S240. doi:10.1093/cid/civ709
80. Jeon HJ, Im J, Haselbeck A, et al. How can the typhoid fever surveillance in Africa and the severe typhoid fever in Africa programs contribute to the introduction of typhoid conjugate vaccines?. Clin Infect Dis. 2019;69(Supplement_6):S417–S421. doi:10.1093/cid/ciz629
81. Walters MS, Routh J, Mikoleit M, et al. Shifts in geographic distribution and antimicrobial resistance during a prolonged typhoid fever outbreak—Bundibugyo and Kasese Districts, Uganda, 2009–2011. PLoS Negl Trop Dis. 2014;8(3):e2726. doi:10.1371/journal.pntd.0002726
82. Amsalu T, Genet C, Adem Siraj Y. Salmonella Typhi and Salmonella Paratyphi prevalence, antimicrobial susceptibility profile and factors associated with enteric fever infection in Bahir Dar, Ethiopia. Sci Rep. 2021;11(1):7359. doi:10.1038/s41598-021-86743-9
83. Awol RN, Reda DY, Gidebo DD. Prevalence of Salmonella enterica serovar Typhi infection, its associated factors and antimicrobial susceptibility patterns among febrile patients at Adare general hospital, Hawassa, southern Ethiopia. BMC Infect Dis. 2021;21(1):1–9. doi:10.1186/s12879-020-05726-9
84. Tack B, Phoba M-F, Van Puyvelde S, et al. Salmonella Typhi from blood cultures in the Democratic Republic of the Congo: a 10-year surveillance. Clin Infect Dis. 2019;68(Supplement_2):S130–S7. doi:10.1093/cid/ciy1116
85. Akinyemi KO, Oyefolu AOB, Mutiu WB, et al. Typhoid fever: tracking the trend in Nigeria. Am J Trop Med Hyg. 2018;99(3_Suppl):41–47. doi:10.4269/ajtmh.18-0045
86. Mashe T, Gudza-Mugabe M, Tarupiwa A, et al. Laboratory characterisation of Salmonella enterica serotype Typhi isolates from Zimbabwe, 2009–2017. BMC Infect Dis. 2019;19(1):1–9. doi:10.1186/s12879-019-4114-0
87. Stanaway JD, Parisi A, Sarkar K, et al. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19(12):1312–1324. doi:10.1016/S1473-3099(19)30418-9
88. Park SE, Pak GD, Aaby P, et al. The relationship between invasive nontyphoidal Salmonella disease, other bacterial bloodstream infections, and malaria in sub-Saharan Africa. Clin Infect Dis. 2016;62(suppl_1):S23–S31. doi:10.1093/cid/civ893
89. Verani JR, Toroitich S, Auko J, et al. Burden of invasive nontyphoidal Salmonella disease in a rural and urban site in Kenya, 2009–2014. Clin Infect Dis. 2015;61(suppl_4):S302–S9. doi:10.1093/cid/civ728
90. Biggs HM, Lester R, Nadjm B, et al. Invasive Salmonella infections in areas of high and low malaria transmission intensity in Tanzania. Clin Infect Dis. 2014;58(5):638–647. doi:10.1093/cid/cit798
91. Nyirenda TS, Nyirenda JT, Tembo DL, et al. Loss of humoral and cellular immunity to invasive nontyphoidal salmonella during current or convalescent Plasmodium falciparum infection in Malawian Children. Clin Vaccine Immunol. 2017;24(7):e00057–17. doi:10.1128/CVI.00057-17
92. Kariuki S, Owusu-Dabo E. Research on invasive nontyphoidal Salmonella disease and developments towards better understanding of epidemiology, management, and control strategies. Clin Infect Dis. 2020;71(Supplement_2):S127–S129. doi:10.1093/cid/ciaa315
93. Uche IV, MacLennan CA, Saul A, Baker S. A systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014). PLoS Negl Trop Dis. 2017;11(1):e0005118. doi:10.1371/journal.pntd.0005118
94. Marchello CS, Fiorino F, Pettini E, et al. Incidence of non-typhoidal Salmonella invasive disease: a systematic review and meta-analysis. J Infect. 2021;83(5):523–532. doi:10.1016/j.jinf.2021.06.029
95. Gilchrist JJ, MacLennan CA, Donnenberg MS, Bäumler AJ. Invasive nontyphoidal salmonella disease in Africa. EcoSal Plus. 2019;8(2). doi:10.1128/ecosalplus.ESP-0007-2018
96. Kariuki S, Mbae C, Onsare R, et al. Multidrug-resistant nontyphoidal salmonella hotspots as targets for vaccine use in management of infections in endemic settings. Clin Infect Dis. 2019;68(Suppl 1):S10–s5. doi:10.1093/cid/ciy898
97. Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56(6):413–422. doi:10.1016/j.jinf.2008.03.012
98. Kingsley RA, Msefula CL, Thomson NR, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19(12):2279–2287. doi:10.1101/gr.091017.109
99. Kariuki S, Revathi G, Kariuki N, et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission?. J Med Microbiol. 2006;55(Pt 5):585–591. doi:10.1099/jmm.0.46375-0
100. Vandenberg O, Nyarukweba DZ, Ndeba PM, et al. Microbiologic and clinical features of Salmonella species isolated from bacteremic children in eastern Democratic Republic of Congo. Pediatr Infect Dis J. 2010;29(6):504–510. doi:10.1097/INF.0b013e3181cd615a
101. Tack B, Phoba M-F, Barbé B, et al. Non-typhoidal Salmonella bloodstream infections in Kisantu, DR Congo: emergence of O5-negative Salmonella Typhimurium and extensive drug resistance. PLoS Negl Trop Dis. 2020;14(4):e0008121. doi:10.1371/journal.pntd.0008121
102. Aldrich C, Hartman H, Feasey N, et al. Emergence of phylogenetically diverse and fluoroquinolone resistant Salmonella Enteritidis as a cause of invasive nontyphoidal Salmonella disease in Ghana. PLoS Negl Trop Dis. 2019;13(6):e0007485. doi:10.1371/journal.pntd.0007485
103. Bachou H, Tylleskär T, Kaddu-Mulindwa DH, et al. Bacteraemia among severely malnourished children infected and uninfected with the human immunodeficiency virus-1 in Kampala, Uganda. BMC Infect Dis. 2006;6(1):160. doi:10.1186/1471-2334-6-160
104. Park SE, Pham DT, Pak GD, et al. The genomic epidemiology of multi-drug resistant invasive non-typhoidal Salmonella in selected sub-Saharan African countries. BMJ Glob Health. 2021;6(8):e005659. doi:10.1136/bmjgh-2021-005659
105. Kariuki S, Okoro C, Kiiru J, et al. Ceftriaxone-resistant Salmonella enterica serotype Typhimurium sequence type 313 from Kenyan patients is associated with the bla CTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob Agents Chemother. 2015;59(6):3133–3139. doi:10.1128/AAC.00078-15
106. Pulford CV, Perez-Sepulveda BM, Canals R, et al. Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa. Nat Microbiol. 2021;6(3):327–338. doi:10.1038/s41564-020-00836-1
107. Falay D, Kuijpers LMF, Phoba M-F, et al. Microbiological, clinical and molecular findings of non-typhoidal Salmonella bloodstream infections associated with malaria, Oriental Province, Democratic Republic of the Congo. BMC Infect Dis. 2016;16(1):1–14. doi:10.1186/s12879-016-1604-1
108. Obaro SK, Hassan-Hanga F, Olateju EK, et al. Salmonella bacteremia among children in central and northwest Nigeria, 2008–2015. Clin Infect Dis. 2015;61(suppl_4):S325–S331. doi:10.1093/cid/civ745
109. Moon HY, Lee DW, Sim GH, et al. A new set of rDNA-NTS-based multiple integrative cassettes for the development of antibiotic-marker-free recombinant yeasts. J Biotechnol. 2016;233:190–199. doi:10.1016/j.jbiotec.2016.07.006
110. Kariuki S, Mbae C, Van Puyvelde S, et al. High relatedness of invasive multi-drug resistant non-typhoidal Salmonella genotypes among patients and asymptomatic carriers in endemic informal settlements in Kenya. PLoS Negl Trop Dis. 2020;14(8):e0008440. doi:10.1371/journal.pntd.0008440
111. Andrews JR, Ryan ET. Diagnostics for invasive Salmonella infections: current challenges and future directions. Vaccine. 2015;33:C8–C15. doi:10.1016/j.vaccine.2015.02.030
112. Kanteh A, Sesay AK, Alikhan N-F, et al. Invasive atypical non-typhoidal Salmonella serovars in The Gambia. Microb Genom. 2021;7(11):000677.
113. Tapia MD, Tennant SM, Bornstein K, et al. Invasive nontyphoidal Salmonella infections among children in Mali, 2002–2014: microbiological and epidemiologic features guide vaccine development. Clin Infect Dis. 2015;61(suppl_4):S332–S338. doi:10.1093/cid/civ729
114. Mandomando I, Bassat Q, Sigaúque B, et al. Invasive Salmonella infections among children from rural Mozambique, 2001–2014. Clin Infect Dis. 2015;61(suppl_4):S339–S345. doi:10.1093/cid/civ712
115. Feasey NA, Masesa C, Jassi C, et al. Three epidemics of invasive multidrug-resistant Salmonella bloodstream infection in Blantyre, Malawi, 1998–2014. Clin Infect Dis. 2015;61(suppl_4):S363–S371. doi:10.1093/cid/civ691
116. Tabu C, Breiman RF, Ochieng B, et al. Differing burden and epidemiology of non-Typhi Salmonella bacteremia in rural and urban Kenya, 2006–2009. PLoS One. 2012;7(2):e31237. doi:10.1371/journal.pone.0031237
117. Okomo U, Akpalu ENK, Le Doare K, et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19(11):1219–1234. doi:10.1016/S1473-3099(19)30414-1
118. WHO. Report on the Burden of Endemic Health Care-Associated Infection Worldwide. WHO; 2011.
119. Scherbaum M, Kösters K, Mürbeth RE, et al. Incidence, pathogens and resistance patterns of nosocomial infections at a rural hospital in Gabon. BMC Infect Dis. 2014;14(1):1–8. doi:10.1186/1471-2334-14-124
120. Shrivastava S, Shrivastava P, Ramasamy J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2018;32(1):76–77. doi:10.4103/jms.jms_25_17
121. Ali S, Birhane M, Bekele S, et al. Healthcare associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: longitudinal study. Antimicrob Resist Infect Control. 2018;7(1):1–9. doi:10.1186/s13756-017-0298-5
122. Sahiledengle B, Seyoum F, Abebe D, et al. Incidence and risk factors for hospital-acquired infection among paediatric patients in a teaching hospital: a prospective study in southeast Ethiopia. BMJ open. 2020;10(12):e037997. doi:10.1136/bmjopen-2020-037997
123. Yallew WW, Kumie A, Yehuala FM, Folgori L. Risk factors for hospital-acquired infections in teaching hospitals of Amhara regional state, Ethiopia: a matched-case control study. PLoS One. 2017;12(7):e0181145. doi:10.1371/journal.pone.0181145
124. Khan HA, Baig FK, Mehboob R. Nosocomial infections: epidemiology, prevention, control and surveillance. Asian Pac J Trop Biomed. 2017;7(5):478–482. doi:10.1016/j.apjtb.2017.01.019
125. Henson SP, Boinett CJ, Ellington MJ, et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307(7):422–429. doi:10.1016/j.ijmm.2017.07.006
126. Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. doi:10.1038/s41579-019-0315-1
127. Cornick J, Musicha P, Peno C, et al.. Genomic investigation of a suspected Klebsiella pneumoniae outbreak in a neonatal care unit in sub-Saharan Africa. Microb Genom. 2021;7(11):000703.
128. Lowe M, Kock MM, Coetzee J, et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg Infect Dis. 2019;25(4):739. doi:10.3201/eid2504.181482
129. Founou RC, Founou LL, Allam M, et al. Whole genome sequencing of extended spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae isolated from hospitalized patients in KwaZulu-Natal, South Africa. Sci Rep. 2019;9(1):1–11. doi:10.1038/s41598-019-42672-2
130. Mbelle NM, Feldman C, Sekyere JO, et al. Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci Rep. 2020;10(1):1–17. doi:10.1038/s41598-020-58012-8
131. Brady MF, Jamal Z, Pervin N. Acinetobacter. In: StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC.; 2022.
132. Ayobami O, Willrich N, Harder T, et al. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg Microbes Infect. 2019;8(1):1747–1759. doi:10.1080/22221751.2019.1698273
133. Villalón P, Ortega M, Sáez-Nieto JA, et al.. Dynamics of a sporadic nosocomial Acinetobacter calcoaceticus – Acinetobacter baumannii complex population. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.00593
134. ECDC. Carbapenem-Resistant Acinetobacter Baumannii in Healthcare Settings– 8 December 2016. Stockholm: ECDC; 2016.
135. Howard A, O’Donoghue M, Feeney A, et al. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence. 2012;3(3):243–250. doi:10.4161/viru.19700
136. Dexter C, Murray GL, Paulsen IT, et al. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther. 2015;13(5):567–573. doi:10.1586/14787210.2015.1025055
137. Montefour K, Frieden J, Hurst S, et al. Acinetobacter baumannii: an emerging multidrug-resistant pathogen in critical care. Crit Care Nurse. 2008;28(1):
138. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi:10.1128/CMR.00058-07
139. Kyriakidis I, Vasileiou E, Pana ZD, et al. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10(3):373. doi:10.3390/pathogens10030373
140. Vázquez-López R, Solano-Gálvez SG, Juárez Vignon-Whaley JJ, et al. Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics. 2020;9(4):205. doi:10.3390/antibiotics9040205
141. Rizk SS, Elwakil WH, Attia AS. Antibiotic-resistant Acinetobacter baumannii in low-income countries (2000–2020): twenty-one years and still below the radar, is it not there or can they not afford to look for it?. Antibiotics. 2021;10(7):764. doi:10.3390/antibiotics10070764
142. Vaara M. Polymyxins and their potential next generation as therapeutic antibiotics. Front Microbiol. 2019;10:1689.
143. Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69(Supplement_7):S521–S528. doi:10.1093/cid/ciz824
144. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
145. Ayibieke A, Kobayashi A, Suzuki M, et al.. Prevalence and Characterization of Carbapenem-Hydrolyzing Class D β-Lactamase-Producing Acinetobacter Isolates From Ghana. Front Microbiol. 2020;11. doi:10.3389/fmicb.2020.587398
146. Odih EE, Irek EO, Obadare TO, et al.. Rectal colonization and nosocomial transmission of carbapenem-resistant Acinetobacter baumannii in an intensive care unit, Southwest Nigeria. Front Med. 2022;9. doi:10.3389/fmed.2022.846051
147. Musila L, Kyany’a C, Maybank R, et al. Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS One. 2021;16(2):e0246937. doi:10.1371/journal.pone.0246937
148. Ssekatawa K, Byarugaba DK, Wampande E, et al. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11(1):629. doi:10.1186/s13104-018-3738-2
149. Olu-Taiwo MA, Opintan JA, Codjoe FS, et al. Metallo-beta-lactamase-producing Acinetobacter spp. from clinical isolates at a tertiary care hospital in Ghana. Biomed Res Int. 2020;2020:3852419. doi:10.1155/2020/3852419
150. Ogbolu DO, Alli OAT, Oluremi AS, et al. Contribution of NDM and OXA-type carbapenemases to carbapenem resistance in clinical Acinetobacter baumannii from Nigeria. Infect Dis. 2020;52(9):644–650. doi:10.1080/23744235.2020.1775881
151. Murray CJL, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655.
152. Ma C, McClean S. Mapping global prevalence of Acinetobacter baumannii and recent vaccine development to tackle it. Vaccines. 2021;9(6):570. doi:10.3390/vaccines9060570
153. Lowe M, Ehlers MM, Ismail F, et al.. Acinetobacter baumannii: epidemiological and beta-lactamase data from two tertiary academic hospitals in Tshwane, South Africa. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.01280
154. Ayenew Z, Tigabu E, Syoum E, et al. Multidrug resistance pattern of Acinetobacter species isolated from clinical specimens referred to the Ethiopian Public Health Institute: 2014 to 2018 trend anaylsis. PLoS One. 2021;16(4):e0250896. doi:10.1371/journal.pone.0250896
155. Aruhomukama D, Najjuka CF, Kajumbula H, et al. blaVIM- and blaOXA-mediated carbapenem resistance among Acinetobacter baumannii and Pseudomonas aeruginosa isolates from the Mulago hospital intensive care unit in Kampala, Uganda. BMC Infect Dis. 2019;19(1):853. doi:10.1186/s12879-019-4510-5
156. Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. doi:10.1016/S1473-3099(18)30605-4
157. Gitau W, Masika M, Musyoki M, et al. Antimicrobial susceptibility pattern of Staphylococcus aureus isolates from clinical specimens at Kenyatta National Hospital. BMC Res Notes. 2018;11(1):226. doi:10.1186/s13104-018-3337-2
158. Ackers-Johnson G, Kibombo D, Kusiima B, et al.. Antibiotic resistance profiles and population structure of disease-associated Staphylococcus aureus infecting patients in Fort Portal Regional Referral Hospital, Western Uganda. Microbiology. 2021;167(5). doi:10.1099/mic.0.001000
159. Egyir B, Guardabassi L, Sørum M, et al. Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from Healthcare Institutions in Ghana. PLoS One. 2014;9(2):e89716. doi:10.1371/journal.pone.0089716
160. Ayepola OO, Olasupo NA, Egwari LO, et al. Molecular characterization and antimicrobial susceptibility of Staphylococcus aureus isolates from clinical infection and asymptomatic carriers in Southwest Nigeria. PLoS One. 2015;10(9):e0137531. doi:10.1371/journal.pone.0137531
161. Perovic O, Iyaloo S, Kularatne R, et al. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2016;10(12):e0145429. doi:10.1371/journal.pone.0145429
162. Verdú-Expósito C, Romanyk J, Cuadros-González J, et al. Study of susceptibility to antibiotics and molecular characterization of high virulence Staphylococcus aureus strains isolated from a rural hospital in Ethiopia. PLoS One. 2020;15(3):e0230031. doi:10.1371/journal.pone.0230031
163. Vubil D, Garrine M, Ruffing U, et al.. Molecular characterization of community acquired Staphylococcus aureus bacteremia in young children in Southern Mozambique, 2001–2009. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.00730
164. Wangai FK, Masika MM, Maritim MC, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in East Africa: red alert or red herring?. BMC Infect Dis. 2019;19(1):596. doi:10.1186/s12879-019-4245-3
165. Shuping LL, Kuonza L, Musekiwa A, et al. Hospital-associated methicillin-resistant Staphylococcus aureus: a cross-sectional analysis of risk factors in South African tertiary public hospitals. PLoS One. 2017;12(11):e0188216. doi:10.1371/journal.pone.0188216
166. Steinhaus N, Al-Talib M, Ive P, et al. The management and outcomes of Staphylococcus aureus bacteraemia at a South African referral hospital: a prospective observational study. Int J Infect Dis. 2018;73:78–84. doi:10.1016/j.ijid.2018.06.004
167. Kateete DP, Namazzi S, Okee M, et al. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes. 2011;4:326. doi:10.1186/1756-0500-4-326
168. Amoako DG, Bester LA, Somboro AM, et al. Plasmid-mediated resistance and virulence mechanisms in the private health sector in KwaZulu-Natal, South Africa: an investigation of methicillin resistant Staphylococcus aureus (MRSA) clinical isolates collected during a three month period. Int J Infect Dis. 2016;46:38–41. doi:10.1016/j.ijid.2016.03.019
169. Donkor ES, Jamrozy D, Mills RO, et al. A genomic infection control study for Staphylococcus aureus in two Ghanaian hospitals. Infect Drug Resist. 2018;11:1757. doi:10.2147/IDR.S167639
170. Nyasinga J, Omuse G, Njenga J, et al. Epidemiology of Staphylococcus aureus infections in Kenya: current state, gaps and opportunities. Open J Med Microbiol. 2020;10:204. doi:10.4236/ojmm.2020.104018
171. Schaumburg F, Alabi AS, Peters G, et al. New epidemiology of Staphylococcus aureus infection in Africa. Clin Microbiol Infect. 2014;20(7):589–596. doi:10.1111/1469-0691.12690
172. Kumwenda P, Adukwu EC, Tabe ES, et al. Prevalence, distribution and antimicrobial susceptibility pattern of bacterial isolates from a tertiary Hospital in Malawi. BMC Infect Dis. 2021;21(1):34. doi:10.1186/s12879-020-05725-w
173. Dame JA, Beylis N, Nuttall J, et al. Pseudomonas aeruginosa bloodstream infection at a tertiary referral hospital for children. BMC Infect Dis. 2020;20(1):729. doi:10.1186/s12879-020-05437-1
174. Raman G, Avendano EE, Chan J, et al. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2018;7(1):79. doi:10.1186/s13756-018-0370-9
175. Cholley P, Ka R, Guyeux C, et al. Population structure of clinical Pseudomonas aeruginosa from West and Central African countries. PLoS One. 2014;9(9):e107008–e. doi:10.1371/journal.pone.0107008
176. Zubair K, Iregbu K. Resistance pattern and detection of metallo‑beta‑lactamase genes in clinical isolates of Pseudomonas aeruginosa in a central Nigeria tertiary hospital. Niger J Clin Pract. 2018;21(2):176–182. doi:10.4103/njcp.njcp_229_17
177. Kateete DP, Nakanjako R, Namugenyi J, et al. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago Hospital in Kampala, Uganda (2007–2009). SpringerPlus. 2016;5(1):1308. doi:10.1186/s40064-016-2986-7
178. Kaluba CK, Samutela MT, Kapesa C, et al. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter species at a large tertiary referral hospital in Lusaka, Zambia. Sci African. 2021;13:e00908. doi:10.1016/j.sciaf.2021.e00908
179. Annear D, Black J, Govender S. Multilocus sequence typing of carbapenem resistant Pseudomonas aeruginosa isolates from patients presenting at Port Elizabeth Hospitals, South Africa. Afr J Infect Dis. 2017;11(2):68–74. doi:10.21010/ajid.v11i2.9
180. Ettu A, Oladapo B, Oduyebo O. Prevalence of carbapenemase production in Pseudomonas aeruginosa isolates causing clinical infections in Lagos University Teaching Hospital, Nigeria. African J Clin Exp Microbiol. 2021;22(4):498–503. doi:10.4314/ajcem.v22i4.10
181. Codjoe FS, Brown CA, Smith TJ, et al. Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS One. 2019;14(9):e0222168. doi:10.1371/journal.pone.0222168
182. Founou RC, Founou LL, Allam M, et al. First report of a clinical multidrug-resistant Pseudomonas aeruginosa ST532 isolate harbouring a ciprofloxacin-modifying enzyme (CrpP) in South Africa. J Glob Antimicrob Resist. 2020;22:145–146. doi:10.1016/j.jgar.2020.05.012
183. Yoon E-J, Jeong SH. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front Microbiol. 2021;12. doi:10.3389/fmicb.2021.614058
184. Kateete DP, Nakanjako R, Okee M, et al. Genotypic diversity among multidrug resistant Pseudomonas aeruginosa and Acinetobacter species at Mulago Hospital in Kampala, Uganda. BMC Res Notes. 2017;10(1):284. doi:10.1186/s13104-017-2612-y
185. Kiyaga S, Kyany’a C, Muraya AW, et al.. Genetic diversity, distribution, and genomic characterization of antibiotic resistance and virulence of clinical Pseudomonas aeruginosa Strains in Kenya. Front Microbiol. 2022;13. doi:10.3389/fmicb.2022.835403
186. Olaniran OB, Adeleke OE, Donia A, et al. Incidence and molecular characterization of carbapenemase genes in association with multidrug-resistant clinical isolates of Pseudomonas aeruginosa from Tertiary Healthcare Facilities in Southwest Nigeria. Curr Microbiol. 2022;79(1):1–14. doi:10.1007/s00284-021-02706-3
187. Philipsborn R, Ahmed SM, Brosi BJ, et al. Climatic drivers of diarrheagenic Escherichia coli incidence: a systematic review and meta-analysis. J Infect Dis. 2016;214(1):6–15. doi:10.1093/infdis/jiw081
188. Aslam B, Khurshid M, Arshad MI, et al.. Antibiotic resistance: one health one world outlook. Front Cell Infect Microbiol. 2021;11. doi:10.3389/fcimb.2021.771510
189. Huijbers PMC, Blaak H, de Jong MCM, et al. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol. 2015;49(20):11993–12004. doi:10.1021/acs.est.5b02566
190. Larsson DGJ, Flach C-F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257–269. doi:10.1038/s41579-021-00649-x
191. Sano D, Louise Wester A, Schmitt H, et al. Updated research agenda for water, sanitation and antimicrobial resistance. J Water Health. 2020;18(6):858–866. doi:10.2166/wh.2020.033
192. Munkholm L, Rubin O. The global governance of antimicrobial resistance: a cross-country study of alignment between the global action plan and national action plans. Global Health. 2020;16(1):109. doi:10.1186/s12992-020-00639-3
193. Elton L, Thomason MJ, Tembo J, et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob Resist Infect Control. 2020;9(1):1–11. doi:10.1186/s13756-020-00800-y
194. Iwu CD, Patrick SM. An insight into the implementation of the global action plan on antimicrobial resistance in the WHO African region: a roadmap for action. Int J Antimicrob Agents. 2021;58(4):106411. doi:10.1016/j.ijantimicag.2021.106411
195. Gordon N, Aggarwal V, Amos B, et al. The UK Fleming Fund: developing AMR surveillance capacity in low-and middle-income countries. Int J Infect Dis. 2020;101:40. doi:10.1016/j.ijid.2020.09.137
196. WHO. Antimicrobial Resistance: National Action Plans. Webpage. WHO; 2016.
197. Mpundu M. Moving from paper to action-the status of national AMR action plans in African countries. Available from: https://revive.gardp.org/moving-from-paper-to-action-the-status-of-national-amr-action-plans-in-african-countries. Accessed July 2, 2022.
198. Goldberg J, Clezy K, Jasovský D, et al. Leaving no one behind: the need for a truly global response to antimicrobial resistance. Lancet Microbe. 2022;3(1):e2–e3. doi:10.1016/S2666-5247(21)00303-7
199. World Health Organization. UHC in Africa: A Framework for Action. World Health Organization; 2016.
200. World Health Organization. The World Health Report: Health Systems Financing: The Path to Universal Coverage: Executive Summary. World Health Organization; 2010.
201. Oleribe OO, Momoh J, Uzochukwu BS, et al. Identifying key challenges facing healthcare systems in Africa and potential solutions. Int J Gen Med. 2019;12:395. doi:10.2147/IJGM.S223882
202. Pierce J, Apisarnthanarak A, Schellack N, et al. Global antimicrobial stewardship with a focus on low-and middle-income countries: a position statement for the international society for infectious diseases. Int J Infect Dis. 2020;96:621–629. doi:10.1016/j.ijid.2020.05.126
203. WHO. WHO Implementation Handbook for National Action Plans on Antimicrobial Resistance: Guidance for the Human Health Sector. WHO; 2022.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.