Back to Journals » Infection and Drug Resistance » Volume 12
Antimicrobial Resistance and Resistance Determinant Insights into Multi-Drug Resistant Gram-Negative Bacteria Isolates from Paediatric Patients in China
Authors Patil S , Chen H, Zhang X , Lian M, Ren PG, Wen F
Received 18 July 2019
Accepted for publication 17 October 2019
Published 22 November 2019 Volume 2019:12 Pages 3625—3634
DOI https://doi.org/10.2147/IDR.S223736
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Sandip Patil,1,2 Hongyu Chen,2 Xiaoli Zhang,2 Ma Lian,2 Pei-Gen Ren,1 Feiqiu Wen2
1Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, Guangdong Province, People’s Republic of China; 2Shenzhen Children’s Hospital, Futian District, Shenzhen, Guangdong Province, People’s Republic of China
Correspondence: Feiqiu Wen
Shenzhen Children’s Hospital, 7019 Yitian Road, Futian District, Shenzhen, Guangdong Province 518038, People’s Republic of China
Tel +86 18938690333
Fax +86-755 83009888
Email [email protected]
Introduction: The emergence of multi-drug-resistant Gram-negative bacteria (GNB) is a concern in China and globally. This study investigated antimicrobial resistance traits and resistance determinant detection in GNB isolates from paediatric patients in China.
Methods: In the present study, a total of 170 isolates of GNB including the most prevalent Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii were collected from Shenzhen Children’s Hospital, China. ESBLs production was confirmed by using the combination disc diffusion method, and carbapenemase production was confirmed by using a carbapenem inactivation method followed by antimicrobial susceptibility. In addition, β-lactamase-encoding genes and co-existence of plasmid-borne colistin resistance mcr-1 gene were determined by PCR and sequencing.
Results: Overall, 170 etiological agents (GNB) were recovered from 158 paediatric patients. The most prevalent species was E. coli 40% (n=68), followed by K. pneumoniae 17.64% (n=30), and Enterobacter cloacae 14.11% (n=24). Of 170 GNB, 71.76% (n=122) were multi-drug-resistant, 12.35% (n=21) extreme-drug resistant, and 7.64% (n=13) single-drug-resistant, while 8.23% (n=14) were sensitive to all of the studied antibiotics. The prevalence of ESBLs and carbapenemase producers were 60% and 17%, respectively. blaCTX-M was the most prevalent resistance gene (59.42%), followed by blaTEM (41.17%), blaSHV (34.270%), blaKPC (34.11%), blaOXA-48 (18.82%) and blaNDM-1 (17.64%).
Conclusion: The present study provides insights into the linkage between the resistance patterns of GNB to commonly used antibiotics and their uses in China. The findings are useful for understanding the genetics of resistance traits and difficulty in tackling of GNB in paediatric patients.
Keywords: Gram-negative bacteria, antimicrobial susceptibility, ESBLs, carbapenemase, molecular characterization
Introduction
The emergence of infectious diseases caused by multi-drug-resistant (MDR) pathogens is a major problem in the community, especially in children.1,2 MDR or extreme drug-resistant (XDR) GNB contributes to global infectious diseases in paediatric patients.3 Recent reports have shown that the rate of resistance in GNB increases periodically worldwide.4 The genomic adaptions: acquisition of resistance determinant by horizontal gene transfer and/or spontaneous mutation in the genome are two major mechanisms that confer resistance against antibiotics in bacteria.5,6 The genomic mutation is responsible for modification in target sequences, overexpression of target, ie, efflux pump and reduced intake of antibiotics, while acquired resistance traits can modify the target post-translationally, inactivate antibiotics by hydrolysis or chemical modification, or may provide alternative metabolic pathways, etc.7 MDR or XDR GNB is more notable in developing countries due to the restricted antibiotics, indiscriminate use of the drugs, poor hygiene, dietary deficiency and poor governing supervision.8,9 However, the antimicrobial resistance problem is still underestimated because of inadequate or ineffective diagnosis in some clinical settings.9,10 Extended spectrum β-lactamases (ESBLs) and carbapenemases are key resistance. These are a group of plasmids-borne, heterogeneous, complex and rapidly evolving enzymes which are capable of hydrolysing penicillin, cephalosporin, aztreonam and monobactams.11,12 According to Bush-Jacoby-Medeiros classification, ESBLs have been classified into three major groups: TEM, SHV and CTX-M, while carbapenemases enzymes encoded by alleles of the blaKPC gene depict one of the five substantial carbapenemase families, others being the VIM, IMP and Delhi Metallo-β-lactamase (MβL) (NDM), and the OXA-48-like oxacillinases.13–15 The β-lactamase production is most commonly seen among GNB including Escherichia coli (E. coli), Klebsiella pneumoniae (K. pneumoniae), Acinetobacter baumannii (A. baumannii) and Pseudomonas aeruginosa (P. aeruginosa).16 In the past few years, high dissemination of ESBLs and carbapenemases-producing GNB were observed worldwide and alarm to developing countries. Children with MDR bacteraemia are more likely to receive inadequate initial antibiotic therapy and have a higher rate of infectious complications and death.17 These observations suggest a critical need for the promotion of antimicrobial stewardship and reduction in unnecessary antibiotic use and avoid the horizontal gene transfer in the paediatric patients.
Methods
Bacterial Isolation and Identification
A total of 170 non-duplicate clinical isolates (GNB) were collected from 158 patients between October 2018 and May 2019 from Shenzhen Children’s Hospital (SCH), China. This hospital is a major children hospital in the southern area of China. A single specimen was isolated from n=146 (85.88%) paediatric patient’s samples, while two specimens were isolated from n=12 (7%) patient’s samples. Among the 170 GNB, 54.12% (n=92) were from male and 45.88% (n=78) were from female; patients' age ranges from ≥4 months to 12 years. The criteria used for inclusion of the isolates in the present study are as follows: first, isolates must be the Gram-negative. Second, the pathogens may link with the community or hospital-associated infections. Bacterial isolates belonging to family Enterobacteriaceae including E. coli, K. pneumoniae, Enterobacter cloacae (E. cloacae), Proteus vulgaris (P. vulgaris) were isolated on MacConkey Agar (Becton Dickinson, USA), Salmonella species were cultivated on deoxycholate citrate agar (Merck, USA), A. baumannii cultivated on CHROMATM Acinetobacter agar (Merck, USA), while Elizabethkingia meningoseptica (E. meningoseptica), Burkholderia cepacia (B. cepacia) cultivated on blood agar, and P. aeruginosa were cultivated on cetrimide agar (Merck, USA). Sets of biochemical tests were performed to identify isolates. The precise phylogenetic identity of all the GNB isolates was further confirmed by 16S rRNA gene sequencing. Bacterial species used in this study were isolated and characterized in biological safety cabinet Class II Type. The origin of the specimens was as follows: urine n=58, sputum n=51, pus n=38, blood n=18, catheter-associated (CA) n=2, and cerebral spinal fluid (CSF) n=3 (S-1).
Phenotypic Detection of ESBLs Production
The combination disc test was done for phenotypic detection of ESBLs production. The test was performed by using a disc of both cefotaxime and ceftazidime, alone and in combination with clavulanic acid. Control strain was selected from the characterized strain collection of our laboratory, while ATCC25922 was used as a negative control strain. The ESBLs production result was analysed according to the Clinical and Laboratory Standards Institute (CLSI) guideline.18
Phenotypic Detection of CRKP
Carbapenemase production was confirmed by using a newly developed Carbapenem Inactivation Method (CIM) which was first delineated in the year 2015.19 To carry out CIM, an antibiotic susceptibility-testing disc of 10-μg meropenem (MEM) was incubated for 2 hrs in an aqueous suspension of a K. pneumoniae. The disc was removed from the suspension and placed onto a Mueller-Hinton agar (MHA) plate seeded with an ATCC25922 indicator organism; followed by overnight incubation, the zone of inhibition was measured to determine whether the MEM has been hydrolysed (growth of the indicator organism under 14 mm area), or still active (>14 mm large zone of inhibition around the disk). Control strain was selected from the characterized strain collection of our laboratory.
Antimicrobial Susceptibility Patterns
Antimicrobial susceptibility was performed by VITEK@2 compact system (Biomerieux-Ref. No.27530/275660) method for 21 antimicrobial agents, namely, amoxicillin, amikacin, aztreonam, ceftazidime, ciprofloxacin, ceftriaxone, colistin, cefuroxime, cefuroxime axetile, cefazolin, ertapenem, cefepime, cefoxitin, imipenem, levofloxacin, nitrofurantoin, ampicillin/sulbactam, trimethoprim/sulfamethoxazole, tigecycline, tobramycin, piperacillin-tazobactam. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guideline18 and colistin susceptibility was determined according to EUCAST.
Detection of Antimicrobial Resistance Encoding Genes
The standard PCR assay was performed to detect the presence of resistance encoding genes: ESBLs encoding genes (blaTEM, blaSHV, blaCTX-M, blaGES, blaCARB, blaPER, blaVEB and blaOXA) using specific primers previously described (Table 1). In addition, carbapenemase genes (blaKPC and blaNDM) and colistin resistance mcr-1 genes were determined by PCR assay and sequencing. The specific primers were used as described in our previous study (Table 1).20–22 The purified PCR products were sequenced commercially (Sangon Biotech-Shanghai, China). DNA Sequences were analysed by NCBI-BLAST program.
 |
Table 1 List of Primers Used in This Study |
Data Analysis and Statistical Tests
Data were double-entered to Epi Data version 3.1 and transferred to SPSS version 20 and Microsoft Excel software for analysis, and the results were presented as tables, pie-charts and graphs. P-values < 0.05 were considered as statistically significant.
Results
Bacterial Confirmation
A total of 170 GNB most commonly E. coli 40% (n=68), K. pneumoniae 17.64% (n=30), E. cloacae 14.11% (n=24), P. aeruginosa 12.94% (n=22), A. baumannii 5.88% (n=10) (Figure 1).
 |
Figure 1 Distribution of isolated Gram-negative pathogens from Shenzhen Children’s Hospital, China. |
Resistance Trends for Commonly used Antibiotics
We summarized the overall resistance trends of commonly used antibiotics against all the isolated GNB over the study period (Figure 2A and B). Overall, ceftriaxone resistance was observed to be the highest 54.11% (n=92). In addition, It has resistance to trimethoprim/sulfamethoxazole 49.41% (n=84), cefuroxime 48.23% (n=82), ampicillin sulbactam 41.76% (n=71), cefazolin 34.70% (n=59), aztreonam 33.52% (n=57), ciprofloxacin 28.82% (n=49), cefoxitin 22.94% (n=39), amoxicillin 19.41% (n=33), ceftazidime 17.64% (n=30), levofloxacin 15.88% (n=27), nitrofurantoin 14% (n=25), cefepime 12.94% (n=22), tobramycin 10.58% (n=18), tigecycline 9.41% (n=16), piperacillin-tazobactam 8.23% (n=14), imipenem and ertapenem 5.29% (n=9) each, amikacin 2.35% (n=4) colistin 1.76% (n=3) (Figure 2A and B). From isolated strains, E. coli has having resistance against ceftriaxone about 54% (n=37), followed by K. pneumoniae 50% (n=15) and E. cloacae 50% (n=12), Salmonella spp. 50% (n=4), P. aeruginosa 45% (n=10), while C. freundii were found sensitive to cephalosporin group of antibiotics. Other side S. typhimurium showed highest resistance for cephalosporin about 63% (n=5) followed by E. cloacae, K. pneumoniae, E. coli about 58% (n=14), 53% (n=16), and 49% (n=33) respectively, Table 2. Conversely, E. coli, K. pneumoniae, and P. aeruginosa isolates were very low resistance for aminoglycoside group (Table 2).
 |
Table 2 Resistance Patterns of Gram-Negative Bacteria for Selected Antibiotics and Antibiotics Classes |
 |
Figure 2 (A) Number of isolates in which resistance is detected for each antibiotic. (B) Detection percentages of resistance to each antibiotic. |
Resistance Patterns for the Different Isolates
Our results reflected that resistance diversity of the pathogens was not observed in any distinct trends. Among 170 GNB, 71.64% (n=122) shown that MDR phenotype followed by 12.35% (n=21) isolates was shown XDR phonotype, 7.64% (n=13) SDR and 8.23% (n=14) MDS phenotype. Form a total of 68 E. coli isolates, 6 (9%), 8 (12%), 41 (60%) and 13 (19%) were multi-drug sensitive (MDS), single-drug resistance (SDR), MDR, XDR, respectively. The overall prevalence of MDR among all isolates was E. cloacae 96% (n=23) followed by K. pneumoniae 80% (n=24), P. aeruginosa 77% (n=17) and A. baumannii 60% (n=6) Table 3. The prevalence of XDR among all the isolates were A. baumannii 20% (n=2), K. pneumoniae 10% (n=3), P. aeruginosa 9% (n=2). The based on the combination disc test result, 61.76% (n=105) ESBLs producers were identified and carbapenem inactivation method test result confirms that 17% (n=30) were carbapenemase producer. A significant association was observed between ESBLs producers and carbapenemase producer (P-value is 0.003642). No significant association was observed between resistance classes (MDS, SDR, MDR, XDR) and isolated GNB isolates (P value is 0.83077).
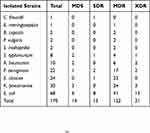 |
Table 3 Distribution of MDS, SDR, MDR, and XDR Pattern of Gram-Negative Bacteria |
Genomics of Resistant Isolates
We explored genome to characterise resistance genes from the GNB. Screening resistance genes showed that Gram-negative isolates harboured blaCTX-M 59.41% (n=101) with most common being blaCTX-M-15 (n=44), blaCTX-M-65 (n=20), blaCTX-M-90 (n-12), blaCTX-M-14 (n=11), blaCTX-M-3 (n=6), blaCTX-M-27 (n=4), blaCTX-M-98, blaCTX-M-211 and blaCTX-M-64 (n=1each) (S-2). Additionally, other β-lactamase encoding genes were detected, blaTEM 41.11% (n=70), blaSHV 34.70% (n=59), blaKPC 34.11% (n=58), blaOXA-48 18.82% (n=32), blaNDM-1 17.64% (n=30), blaGES 14.11% (n=24), blaVIM 9.41% (n=16), blaCARB 8.23% (n=14) most recently discovered plasmid-borne colistin resistance mcr-1 1.76% (n=3) Table 4. The blaPER, blaVEB and blaSME genes were not detected. The significant co-associations were observed between blaCTX-M to blaTNDM-1, blaKPC, and blaTEM (p = 0.003462, 0.00001 and 0.000335, respectively).
 |
Table 4 Prevalence of Drug Resistance Determinant (Genes) in Isolated Gram-Negative Bacteria |
Discussion
Infection caused by GNB is one of the major burdens on low and middle-income countries because of acquisition of resistance genotype. GNB, more precisely, E. coli, K. pneumoniae, E. cloacae, P. vulgari, Salmonella species, account for the most severe forms of infections including urinary tract infection (UTI), bacteraemia and pneumonia. The usual therapeutic approaches to treat such conditions are by antibiotics.24,25 Recent findings on antimicrobial resistance revealed that resistances to frontline antimicrobials among GNB are very common and the resistance can spread from multiple sources through a number of pathogenic or commensal microbes by horizontal gene transfer.26 The overall rate of MDR and XDR of the GNB from SCH were found to be 71% and 12.0%, respectively. Furthermore, the observed MDR rate is significantly associated with prolonged hospital stay and the patients, who die due to MDR bacterial species (even if, it is not statistically significant). On the other hand, the observed XDR rate at our hospital indicates that the problem of antimicrobial resistance is increasing at an alarming rate and circulating Gram-negative pathogenic bacteria in SCH are becoming more resistant to available all available antibiotics. Recent reports from Wenzhou, China have indicated that 60.1% MDR gram-negative pathogen infection in the children’s hospital is almost similar to our results.27 The occurrence of a high rate of MDR gram-negative pathogenic bacteria would also have huge potential threat and implications for children’s care in the hospital and the community at large. As we are living in the Shenzhen (International City) of a very connected world, it is highly likely for these MDR bacteria to be disseminated to other parts of China and globally.
To the best of our knowledge, there is no previous report from Shenzhen on the rate of MDR and XDR gram-negative pathogenic bacteria from paediatric patients and genetic resistance determinant analysis to compare with these results. The antimicrobial susceptibility test data clearly indicate that a high resistance rate of the gram-negative pathogen to the cephalosporin group of antibiotics has raised a serious concern and become a challenge for clinicians. Therefore, we suggest avoiding indiscriminate use of antibiotics in medical practice which will certainly lower the opportunities for the emergence of resistance. Our antimicrobial susceptibility results are comparable to another part of China, Taiwan, and the USA.28,29 The ESBLs detection test confirms 59% (n=101) ESBLs producers, while 17% were carbapenemase producer which is quite higher than our previous study,17 but comparable to studies done in Nigeria, Nepal and India.30,31 Development of AMR is an outcome of complex microbial interactions and resistance may arise by the acquisition of de-novo mutation during clinical antibiotic treatment or commonly by the acquisition of integrative or replicative mobile genetic elements (MGEs) that have evolved over time in microbes in the natural ecosystem. The environmental reservoirs of resistance genes are widely diverse and similar resistance genes may present in distantly related bacterial species, which can co-exist in the same habitat.32,33 It was reported that K. pneumoniae, V. cholerae, E. coli, P. aeruginosa, and Salmonella were naturally competent and can uptake naked DNA from the environment in suitable conditions.34,35 In this study, we observed that blaCTX-M as the most prevalent genotype of ESBLs-producing gram-negative pathogens in SCH, which is composed of blaCTX-M-15 followed by blaCTX-M-65, blaCTX-M-90, blaCTX-M-14, blaCTX-M-3, blaCTX-M-27, blaCTX-M-98, blaCTX-M-211 and blaCTX-M-64 (S-2). This result indicates the diversity of CTX-M genotype of ESBLs-producing gram-negative pathogens spread in Shenzhen, China. Additional, blaTEM followed by blaSHV, blaOXA-48, blaGES, blaCARB and blaVIM genes exists in isolated GNB. Similar results were reported from Japan and Tanzania.36,37 In our study, blaCTX-M-55 is not detected normally in pediatric patients, which means children may not be in contact with an animal since this genotype is mostly circulated via animal origin E. coli isolates.38 We do not observe blaVEB, blaPER and blaSME from same isolates. We observed that blaKPC 34% and blaNDM-1 17.64% as the most prevalent carbapenemases encoding genes in GNB. In addition, co-existence of ESBLs encoding blaCTX-M genes with carbapenemases encoding genes blaKPC or blaNDM-1 raises a concern about the spread of superbugs in the Shenzhen. Several reports mentioned the co-existence of blaCTX-M with blaKPC or blaNDM-1 globally.39 We observed three isolates harbouring plasmid-borne colistin resistance mcr-1, even highly spreading over tChina.40 The overall study suggested that limited options are available to treat MDR GNB infection in children.
Conclusion
In past few years, a number of studies have been repeated globally on antimicrobial resistance focused on pathogenicity, prevalence, resistance mechanisms (acquisition and dissemination of resistance genes), as well as on drug-susceptibility testing, rapid diagnosis of AMR pathogens and developing a strategy to re-sensitize the resistance variants against existing drugs of public-health importance. Despite the availability of the plethora of information on the fundamental science of resistance biology, the information on the rise of resistant pathogens across the globe is surprisingly scarce. Our data show that the clinical isolates (gram-negative pathogen) are continuously evolving to counterbalance the bactericidal/bacteriostatic effects of clinically important antimicrobial drugs because of acquisition of resistance elements such as blaCTX-M, blaKPC, blaNDM-1, blaTEM, blaSHV, blaOXA-48, blaGES, blaVIM and blaCARB. It is a right time to develop strategies for robust surveillance, restriction on improper antibiotic usage and identify the routes that are facilitating the rapid dissemination of antibiotic resistance in pathogenic and non-pathogenic bacterial cells in children’s hospitals.
Ethics Statement
The present study was approved by the ethical committee of the Shenzhen Children’s Hospital (Research) 2018 (013). The clinical isolates used in this research were part of routine hospital laboratory procedures. Verbal consent was given by the patient’s parent/s or legal guardian/s.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kate EJ, Nikkita GP, Marc AL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi:10.1038/nature06536
2. Dawood Y, Tamara J, Sajeda K, Rawan AR, Wasim K, Miral A. Community-acquired serious bacterial infections in the first 90 days of life: a revisit in the era of multi-drug-resistant organisms. World J Pediatr. 2019. doi:10.1007/s12519-019-00276-w
3. Spyridon K, Hamid B, George S, Michael M, Constantinos T. Intravenous colistin use for infections due to MDR GNB in critically ill paediatric patients: a systematic review and meta-analysis. J Antimicrob Chemother. 2019. print ahead. doi:doi:10.1093/jac/dkz165.
4. Martin E, Sanjay B, Bärbel C, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:1–24. doi:10.3205/dgkh000290
5. Baym M, Stone K, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:aad3292. doi:10.1126/science.aad3292
6. Alison H, Luke P, Arnfinn S, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2015;387(100140):176–187. doi:10.1016/S01406736(15)00473-0.
7. Étienne R, Paul-Louis W, François B. Mechanisms of antimicrobial resistance in GNB. Ann Intensive Care. 2015;5:21. doi:doi:10.1186/s13613-015-0061-0
8. Colodner R, Rock W, Chazan B, et al. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in non-hospitalized patients. Eur J Clin Microbiol Infect Dis. 2004;23(3):163–167. doi:10.1007/s10096-003-1084–2
9. Ayukekbong A, Ntemgwa M, Atabe N. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):47. doi:10.1186/s13756-017-0208-x.
10. World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; 2014.
11. Fernando M, Luke W, Miththinda J, et al. Extended spectrum beta-lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern – a hospital based cross sectional study. BMC Infect Dis. 2017;17(1):138. doi:10.1186/s12879-017-2250-y.
12. Pitout D, Laupland B. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–166. doi:10.1016/S1473-3099(08)70041-0
13. Queenan M, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi:doi:10.1128/CMR.00001-07
14. Chang-Ro L, Jung L, Kwang P, Young K, Byeong J, Sang L. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi:10.3389/fmicb.2016.00895
15. Stoesser N, Sheppard E, Peirano G, et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep. 2017;7(1):5917. doi:10.1038/s41598-017-06256-2
16. Mao T, Zhai H, Duan G, Yang H. Patterns of drug-resistant bacteria in a General Hospital, China, 2011–2016. Pol J Microbiol. 2019;68(2):225–232. doi:doi:10.33073/pjm-2019-024.
17. Sandip P, Xiaowen C, Ma L, Feiqiu W. Phenotypic and genotypic characterization of multi-drug-resistant Escherichia coli isolates harbouring blaCTX-M group extended-spectrum β-lactamases recovered from pediatric patients in Shenzhen, southern China. Infect and Drug Resist. 2019;12:1325–1332. doi:10.2147/IDR.S199861
18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth Informational Supplement; 2010. Available from: https://clsi.org/.
19. Kim Z, Angela H, Gerlinde P, Hester B, Albert N, Leo S. The Carbapenem Inactivation Method (CIM), a simple and low-cost alternative for the carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One. 2015;10(3):e0123690. doi:doi:10.1371/journal.pone.0123690
20. Yi-Yun L, Yang W, Timothy W, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:doi:10.1016/S1473-3099(15)00424-7
21. Yee-Huang K, Mei-Feng L, Yin-Ching C, Wen-Liang Y. Detection of plasmid-mediated β-Lactamase genes and emergence of a novel AmpC (CMH-1) in Enterobacter cloacae at a medical Center in Southern Taiwan. J Clin Med. 2019;8(1):8. doi:10.3390/jcm8010008
22. Tian B, Huang M, Fang L, Qing Y, Zhang F, Huang X. CTX-M-137, a hybrid of CTX-M-14-like and CTX-M-15-like beta-lactamases identified in an Escherichia coli clinical isolate. J Antimicrob Chemother. 2014;69(8):2081–2085. doi:10.1093/jac/dku126
23. Magiorakos AP, Srinivasan A, Carey R, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
24. Hye Sun H, Ji Hye K, Myung Hyun C, Eujin P. Low relapse rate of urinary tract infections from extended-spectrum beta-lactamase-producing bacteria in young children. Pedi Nephro. 2019. in press. doi:doi:10.1007/s00467-019-04298-4.
25. Khalili H, Afhami S, Dashti-Khavidaki S, Alijani B. Antimicrobial resistance pattern of Gram-negative bacteria of nosocomial origin at a teaching hospital in the Islamic Republic of Iran. Eastern Medi Terranean Health J. 2012;18(2):172–177. doi:10.26719/2012.18.2.172
26. Woolhouse ME, Ward MJ. Sources of antimicrobial resistance. Science. 2013;341(6153):1460–1461. doi:10.1126/science.1243444.
27. Dong L, Zhang XY, Li CC, Li Z, Xia YQ. Characteristics of epidemiology and antimicrobial resistance of gram-negative bacterial bloodstream infections in children. Chin J Paediatr. 2017;55(9):683–688. doi:10.3760/cma.j.issn.0578-1310.2017.09.012.
28. Gales AC, Castanheira M, Jones RN, Sader HS. Antimicrobial resistance among GNB isolated from Latin America: results from SENTRY antimicrobial surveillance program (Latin America, 2008–2010). Diagn Microbiol Infect Dis. 2012;73(4):354–360. doi:doi:10.1016/j.diagmicrobio.2012.04.007
29. Fei H, Wei-Yang L, Jiun-Ling W, et al. Faecal carriage of multidrug-resistant Escherichia coli by community children in southern Taiwan. BMC Gastroenterol. 2018;18(86):1–8. doi:doi:10.1186/s12876-017-0727-1
30. Parajuli NP, Acharya SP, Mishra SK, Parajuli K, Rijal BP, Pokhrel BM. High burden of antimicrobial resistance among gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob Resist Infect Control. 2017;6(1):67. doi:10.1186/s13756-017-0222-z.
31. Shukla I, Tiwari R, Agrawal M. Prevalence of extended spectrum-lactamase producing Klebsiella pneumoniae in a tertiary care hospital. Indian J Med Microbiol. 2004;22(2):87.
32. Wellington EM, Boxall AB, Cross P, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013;13(2):155–165. doi:10.1016/S1473-3099(12)70317-1
33. Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2):1–37. doi:10.1128/microbiolspec.VMBF-0016–2015
34. Hughes D. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat Rev Genet. 2003;4(6):432–441. doi:10.1038/nrg1084(2003)
35. Gelband H, Laxminarayan R. Tackling antimicrobial resistance at global and local scales. Trends Microbiol. 2015;23(9):524–526. doi:10.1016/j.tim.2015.06.005
36. Sonda T, Kumburu H, Van Zwetselaar M, et al. Prevalence and risk factors for CTX-M gram-negative bacteria in hospitalized patients at a tertiary care hospital in Kilimanjaro, Tanzania. Eur J Clin Microbiol Infect Dis. 2018;37(5):897–906. doi:10.1007/s10096-018-3196-8.
37. Shin-ichi F, Kentaro Y, Thikako O, Kouhei U, Yukiko T, Yasuko S. Rapid identification of gram-negative bacteria with and without CTX-M extended-spectrum β-Lactamase from positive blood culture bottles by PCR Followed by microchip gel electrophoresis. J Clin Microbiol. 2011;49(4):1483–1488. doi:doi:10.1128/JCM.01976–10
38. Li S, Zhao M, Liu J, Zhou Y, Miao Z. Prevalence and antibiotic resistance profiles of extended-spectrum β-lactamase-producing isolated from healthy broilers in Shandong province, China. J Food Prot. 2016;79(7):1169–1173. doi:doi:10.4315/0362-028X.JFP-16-025.
39. Ma J, Liu J-H, Lv L, et al. Characterization of extended-spectrum β-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl Environ Microbiol. 2012;78(10):3668–3673. doi:doi:10.1128/AEM.07507-11.
40. Ruobing W, Lucy VD, Liam P, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9:1179. doi:10.1038/s41467-018-03205-z
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
