Back to Journals » Infection and Drug Resistance » Volume 12
Antimicrobial Resistance and Molecular Characteristics of Methicillin-resistant Staphylococcus aureus Isolates from Children Patients in Iran
Authors Samadi R , Ghalavand Z, Mirnejad R, Nikmanesh B, Eslami G
Received 31 August 2019
Accepted for publication 11 November 2019
Published 9 December 2019 Volume 2019:12 Pages 3849—3857
DOI https://doi.org/10.2147/IDR.S229394
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Roghayeh Samadi,1 Zohreh Ghalavand,1 Reza Mirnejad,2 Bahram Nikmanesh,3 Gita Eslami1
1Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Molecular Biology Research Center, Systems Biology and Poisoning Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran; 3Department of Medical Laboratory Sciences, School of Allied Medical Sciences, Tehran University of Medical Sciences, Tehran, Iran
Correspondence: Gita Eslami
Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Tel +98-2123872556
Email [email protected]
Introduction: Methicillin-resistant Staphylococcus aureus (MRSA) causes high rates of mortality and a substantial burden to health systems worldwide. Here, we investigated the antimicrobial susceptibility and molecular characteristics of MRSA isolated from children referred to Children’s Medical Center in Tehran.
Materials and methods: A total of 98 MRSA isolates were collected from children. Antimicrobial resistance patterns were determined using the disk diffusion and E-test methods. The presence of biofilm encoding genes and the pvl gene were determined by PCR. We used the microtiter plate method to assess the ability of biofilm formation. The MRSA isolates were further analyzed using PFGE and SCCmec typing.
Results: Antibiotic susceptibility testing showed that the highest and the lowest antibiotic resistance percentage were related to erythromycin (62%) and minocycline (10%), respectively. Overall, 63% of MRSA isolates were biofilm producers. Resistance to two antibiotics such as erythromycin (72% vs 28%, P=0.01) and clindamycin (71% vs 29%, P=0.04) was higher among biofilm producers than non-biofilm producers. All strains had biofilm-forming genes and the prevalence of pvl gene was 41%. Most MRSA isolates belonged to SCCmec IVa (75%) and SCCmec III (18%). In PFGE technique, 5 common types and 2 single types were identified; Common type 1 with 37 isolates was dominant clone.
Conclusion: We thus report preliminary data on the prevalence and distribution of MRSA genotypes in Tehran Children’s Hospital. These findings characterize the MRSA colonization dynamics in child patients in Iran and may aid the design of strategies to prevent MRSA infection and dissemination.
Keywords: MRSA, biofilm formation, antibiotic resistance, SCCmec typing, PFGE
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a common pathogen causing various forms of infectious disease in humans.1 Children colonized with MRSA are potential reservoirs for the spread of MRSA in the community.2 Furthermore, immunologically immature infants and newborns, especially those born prematurely or requiring specialized care, are most susceptible to MRSA infections.3
MRSA biofilm formation is regulated by the expression of polysaccharide intracellular adhesion (PIA), which mediates cell to cell adhesion and is encoded by the icaADBC operon.4 Moreover, surface-associated proteinaceous adhesins can contribute to the adherence, colonization and biofilm formation of MRSA. This pathogen can express a variety of microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), such as fibronectin-binding proteins A and B (FnbA, FnbB), clumping factors A and B (ClfA, ClfB), collagen-binding protein (Cna) and enolase protein (eno). Biofilm formation interferes with bacterial recognition and killing mechanisms of the innate immune system.5,6
A number of methods have been created for the detection of biofilm formation ability. Currently, several different methods are used, such as tube test, microtiter plate test, radiolabeling, microscopy and Congo red agar plate test (CRA).7,8 However, the microtiter plate method (Mtp) is a quantitative and reliable method to detect biofilm-forming bacteria. Compared to tube and CRA methods, it can be recommended as a general screening method for the detection of biofilm-producing bacteria in laboratories.9,10
Molecular typing methods have been applied to help researchers map the spread and evolution of MRSA clones, including pulsed-field gel electrophoresis (PFGE) and staphylococcal cassette chromosome mec typing (SCCmec typing).11,12
PFGE is still considered a standard reference molecular technique for analyzing the dissemination of hospital and community-acquired MRSA and has proved to be one of the most discriminatory methods in the total sequencing of the MRSA strain.13 It has been an excellent laboratory tool for emergency identification of new clones.14
Staphylococcal cassette chromosome mec (SCCmec) typing accompanied with overall genotyping has already provided strong evidence for the independent origins of health-care associated MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA).15 To date, eight different types of SCCmec (I–VIII) have been defined on the basis of the combination of ccr and mec complexes, but only types I–V are globally distributed, while others appear to exist as local strains in the country of origin.16–18 PVL is a two-component S. auerus spore-forming protein encoded by the lukF-PV and lukS-PV genes.19 PVL toxin is responsible for the increased virulence of CA-MRSA, since the gene is responsible for many of the severe clinical syndromes of MRSA such as severe necrotizing pneumonia.20,21
However, epidemiological analysis among clinical MRSA isolates from children has rarely been performed. The aim of this study was to investigate the antimicrobial resistance pattern, biofilm formation and molecular characteristics of MRSA strains in children.
Materials and Methods
Bacterial Strains
In this cross-sectional study, 98 suspected staphylococcal infection samples were routinely collected from patients referred to the pediatric medical center and then specimens infected with Methicillin-resistant S. aureus were included in our study for a specified period (from September 2016 to October 2017). S. aureus isolates were confirmed using conventional microbiological methods (Gram’s stain, catalase, coagulase, DNase tests and mannitol fermentation on mannitol salt agar (Merck, Germany)). To definitively identify positive S. aureus isolates, they were subjected to polymerase chain reaction (PCR) for nucA gene. MRSA strains were identified phenotypically using cefoxitin disk-diffusion method (30 μg; MAST, UK). This method was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.22 Resistance to methicillin in S. aureus isolates was confirmed by the amplification of mec gene by PCR method.
Antimicrobial Susceptibility Testing of MRSA Isolates
The antibiotic susceptibility patterns of MRSA isolates were determined by the Kirby-Bauer disk-diffusion method, and the results were interpreted according to CLSI guidelines.22 The antimicrobial agents (Rosco, Denmark) tested in this study included clindamycin (2 μg), linezolid (30 μg), penicillin (10 μg), gentamicin (10 μg), trimethoprim-sulfamethoxazole (25 μg), minocycline (30 μg) and erythromycin (15 μg). S. aureus ATCC 25923 was used as a standard strain. The minimum inhibitory concentration (MIC) for vancomycin was determined with E-test strips (Liofilchem, Italy) according to manufacturer's instructions. The standard reference strain S. aureus ATCC 25923 was used as a quality control strain in every test run.
Detection of Biofilm Formation by Microtiter Plate Assay (Mtp)
Biofilm production was determined quantitatively using microtiter plate method as described previously.23 Briefly, bacterial isolates were grown in Brain Heart Infusion (BHI) with 1% glucose (Merck, Germany) and incubated at 37ºC for overnight.24 Cultures were diluted 1:20 in fresh BHI-0.1% glucose. Then, 200 μL of the diluted solution was added to wells of a flat-bottomed polystyrene microtitre plate and incubated for 48 hrs at 37ºC. The negative control wells contained 200 μL of BHI-0.1% glucose. Wells were gently washed 3 times with phosphate-buffered saline (PBS; pH 7.2), fixed with sodium acetate (2%) for 10 mins, dried at room temperature and then strained with 0.1% crystal violet. After removing the crystal violet solution, wells were washed with PBS to remove unbound dye. The optical densities (ODs) of the plates were observed at 630 nm using a microtiter plate reader. Each assay was performed in duplicate. As a negative control, brain heart infusion broth with 1% glucose medium was used to determine the background OD. OD cut-off was then determined as an average OD of negative control + 3× standard deviation of negative control. The OD cut-off value was separately calculated for each microtiter plate. Biofilm formation by isolates was calculated and categorized according to the absorbance of the crystal violet-stained attached cells (Table 1). Staphylococcus epidermidis ATCC 35984 was used as the biofilm producer control strain.25,26
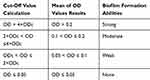 |
Table 1 Classification of Biofilm Formation Abilities by Microtiter Plate Method |
Extraction of Genomic DNA
Genomic DNA was extracted from pure cultures using the High Pure PCR Template Preparation Kit (Roche, Germany), according to the manufacturer’s guidelines. The concentration of DNA was assessed using a spectrophotometer.
Detection of Biofilm Encoding Genes and pvl Gene
All 98 MRSA isolates were tested for the presence of pvl gene and biofilm encoding genes (icaA, icaD, fnbA, fnbB, clfA, clfB, cna, eno) with the degenerate primers as listed in Table 2.
 |
Table 2 Oligonucleotide Primers Used in This Study |
SCCmec Typing
SCCmec typing was performed for all MRSA isolates by multiplex PCR according to the works of Zhang et al.27 Primer sequences are showed in Table 1. SCCmec types were identified by comparing the banding patterns of MRSA to ATCC 10442 (SCCmec type I), N315 (SCCmec type II), 85/2082 (SCCmec type III), CA05 (SCCmec type IVa), 8/6-3P (SCCmec type IVb), MR108 (SCCmec type IVc), JCSC4469 (SCCmec type IVd) and JCSC3624 (SCCmec type V) as reference strains.
Pulsed-Field Gel Electrophoresis
PFGE based on SmaI macrorestriction analysis was performed using the CDC laboratory protocol for S. aureus.28 The PFGE was run on a CHEF DR III system (Bio-Rad, CA, USA) with optimum settings as follows: initial switch 5 s, final switch 40 s, run time 21 hrs, voltage 6 V/cm and a SeaKem Gold agarose (Lonza, Rockland, USA) gel concentration of 1%. Analysis of PFGE clusters was performed using the BioNumerics software package (Applied Maths, Sint-Martens-Latem, Belgium), using the Dice coefficient, and visualized as a dendrogram by the unweighted pair group method.
Statistical Analysis
The relationship between biofilm formation and antibiotic resistance among MRSA isolates was evaluated by the Pearson Chi-Square test using SPSS version 21. P-values less than 0.05 were considered to be significant.
Results
A total of 98 MRSA isolates were collected from children referred to pediatric hospital during a 2014–2015 years. Of these patients, 51 (52%) were girls and 47 (48%) were boys. The median age of the patients was 45 ± 5 months (1 month to 14 years). The MRSA isolates were recovered from respiratory secretions (57%), blood (15%), wounds (10%), the ear (8%), as skin abscesses (5%), the eye (5%). All isolates were susceptible to linezolid and vancomycin and resistant to penicillin and cefoxitin. The rates of resistance to the majority of antibiotics tested varied from 10% to 62% (Figure 1). Linezolid and vancomycin showed good activity against MRSA isolates. The rate of the susceptibility of vancomycin is shown in Table 3.
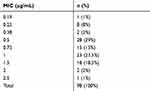 |
Table 3 The MIC Range of Vancomycin in MRSA Isolates |
The results of Mtp assay of MRSA strains are presented in Table 4. Of the 98 MRSA isolates, 62 (63%) strains were biofilm producers. Statistical analysis showed a significant relationship between biofilm formation and antibiotic resistance. Resistance to two antibiotics such as erythromycin (72% vs 28%, P=0.01) and clindamycin (71% vs 29%, P=0.04) was higher among biofilm producers than non-biofilm producers (Table 5). All MRSA strains had biofilm-forming genes and the prevalence of pvl gene was 41%.
 |
Table 4 Percentage of Biofilm Formation in Microtiter Plate Method |
 |
Table 5 Relationship Between Biofilm Formation and Antibiotic Resistance |
Total distribution of SCCmec types in the MRSA strains of various clinical infections is shown in Table 6. Our results showed that the most commonly detected SCCmec types in the MRSA strains were SCCmec IVa (74 strains, 75%) and SCCmec III (18 strains, 18%).
 |
Table 6 Multiplex PCR Results of SCCmec Typing |
PFGE of 98 MRSA strains generated 7 pulsotypes, consisted of five common types (CT1–CT5) and 2 single types (ST1 and ST2). The majority of the strains (38%) were CT1, followed by CT2 which contained 27 strains (28%), CT3 which contained 14 strains (14%), CT4 which contained 12 strains (12%) and CT5 which contained 6 strains (6%) (Figure 2).
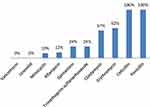 |
Figure 1 Antimicrobial resistance patterns of MRSA isolates. |
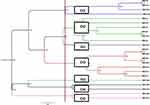 |
Figure 2 Dendrogram of MRSA patterns generated by an UPGMA clustering method. |
Discussion
S. aureus is included in the group of “ESKAPE” bacteria, which comprise the MDR pathogens that are currently considered as the biggest concern for humanity.34,35 There is a relative abundance of the different antibiotic-groups for the treatment of MRSA.36,37 This is underlined by the recent WHO report, urging drug companies to invest and target various drug-resistant bacteria during antibiotics research, which also includes MRSA.38 The pathogenicity of S. aureus is related to the ability to produce toxins and extracellular factors such as biofilms that enable the bacterial adhesion and resistance to phagocytosis.39,40 It is now estimated that biofilms are responsible for more than 65% of nosocomial infections and 80% of all microbial infections.41 In biofilm formation of different bacterial species, the transmission of antimicrobial resistance markers occurs more frequently, and the transfer of antibiotic resistance from Enterococcus to more pathogenic bacteria such as. aureus is a major threat.41 In this study, 63% of isolates were capable of biofilm formation by microtiter plate method, of which 1% were strongly adherent, 8% moderately adherent, 54% weakly adherent and 37% non-adherent, which were matched with the researches conducted by Lotfi et al and Yousefi et al.25,42 Studies show that the microtiter plate method is more sensitive and specific than other methods and has been introduced as a gold standard in biofilm identification.10,23,24 In this study, all isolates were susceptible to vancomycin and linezolid, while (62%) isolates showed resistance to erythromycin, (57%) to clindamycin, (24%) to trimethoprim-sulfamethoxazole, (24%) to gentamicin, (12%) to rifampin and (10%) to minocycline. Although antibiotic resistance was higher in biofilm-producing strains than other strains, there was a statistically significant relationship between antibiotic resistance of erythromycin and clindamycin and biofilm formation. All genes involved in biofilm formation including clfA, clfB, fnbA, fnbB, cna, eno, icaD, icaA were identified in all S. aureus isolates. In the study by Yousefi et al in Iran, the prevalence of biofilm-related genes was 100%,42 while in the study by Mohamed et al in Iraq, the prevalence of fnbA, clfA and cna genes was, respectively, 56%, 56% and 81%.43 The results of this study and other studies indicate that biofilm formation in Staphylococcus strains is dependent on environmental conditions and is influenced by environmental signals that can respond to external stress and inhibitory concentrations of antibiotics.24 Failure in biofilm formation despite the presence of ica genes can be due to the inactivation of ica operons by activation of icaR repressor.44
In the present study, frequency of SCCmec typing was, respectively, SCCmec I (1%), SCCmec III (18%), SCCmec IVa (75%), SCCmecIVb (4%) and SCCmec IVc (2%). SCCmec II, SCCmec IVd and SCCmec V types were not detected. Results of studies indicated that strains carrying large chromosomal cassettes, such as SCCmec I-III, are often resistant to non-β-lactam antibiotic classes and rarely carry the pvl gene. In contrast, strains carrying smaller chromosomal cassettes, such as SCCmec IV and SCCmec V, are less resistant to non-β-lactam antibiotic classes and often carry the pvl gene.45,46 In this study, the strains showed less resistance to non-β-lactam drugs such as gentamicin, minocycline and rifampin, and 41% of the strains carried the pvl gene.
In this study, PFGE technique was used as a powerful discriminative tool to investigate the epidemiological characteristics of MRSA strains. This method has high resolution and reproducibility and is used as the gold standard method for typing this genus. Genotyping techniques such as PFGE are helpful in finding a transferable clone and infection control and prevention measures.
In this study, 5 common types with 4 to 37 subtypes and 2 single types were identified. Common type 1 with 37 isolates was dominant clone and all strains had SCCmec IV, most of which were isolated from outpatients with respiratory infections. Whereas Common type 2 with 27 isolates was often obtained from inpatients in different departments of the hospital. Based on these results, it is likely that Common type 1 colonizes in the respiratory tract of children and circulating in the community, whereas Common type 2 is circulating in hospitals and in different parts of it. Common type 3 with 14 isolates also had the same antibiotic resistance pattern and were only separated from the emergency and surgical departments. A similar study by Ohadian moghadam et al, in 2017, was performed to classify MRSA strains using the PFGE technique in Iran. In this study, 43 MRSA strains were isolated from wound swabs of patients referred to Shahid Motahhari Hospital (specializing in the treatment of burns). After performing PFGE, 5 common types and 31 single types were identified. The investigation indicated that each common type represents an outbreak, because it was taken over an identical time interval and the diversity of strains had been explained by the acquisition of MRSA from various sources.47 Another study by Hussein et al investigated the typing of 114 strains of S. aureus isolated from healthcare workers using the PFGE technique in Iraq. In this study, 8 common types were isolated, more than 50% of isolates belonged to types A and B, indicating infection with the same source.48
In this study, since no sampling of hospital personnel and equipment was carried out, it is impossible to investigate the source of infection and its transmission to patients in different departments of the hospital, which requires a large-scale study, but the important point that should be noted is the infection is transmitted from the community to the hospital, which must be prevented by appropriate infection control measures.
Conclusion
In this study, most of the strains belonged to CA-MRSA because they were mostly carriers of the SCCmec IVa gene and were highly sensitive to non-beta-lactam drugs such as minocycline and rifampin. According to the PFGE technique, cross-sectional circulation of clones was observed in the hospital, which requires careful control of infection in different parts of the hospital.
Acknowledgments
This project was supported financially by the research deputy of Shahid Beheshti University of Medical Sciences.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279(8):593–598. doi:10.1001/jama.279.8.593
2. Wertheim HF, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–762. doi:10.1016/S1473-3099(05)70295-4
3. Ben Nejma M, Merghni A, Mastouri M. Genotyping of methicillin resistant Staphylococcus aureus strains isolated from hospitalized children. Int J Pediatr. 2014;2014.
4. Kaur DC, Wankhede S. Biofilm formation and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus from wound infection. Asian Pac J Health Sci. 2014;1(4):322–328. doi:10.21276/apjhs
5. Tang J, Chen J, Li H, Zeng P, Li J. Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among Staphylococcus aureus strains isolated from different sources. Foodborne Pathog Dis. 2013;10(9):757–763. doi:10.1089/fpd.2012.1474
6. Eyoh AB, Toukam M, Atashili J, et al. Relationship between multiple drug resistance and biofilm formation in Staphylococcus aureus isolated from medical and non-medical personnel in Yaounde, Cameroon. Pan Afr Med J. 2014;17.
7. Deighton MA, Capstick J, Domalewski E, Van Nguyen T. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 2001;336:177–195.
8. Arciola CR, Campoccia D, Gamberini S, Cervellati M, Donati E, Montanaro L. Detection of slime production by means of an optimised Congo red agar plate test based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials. 2002;23(21):4233–4239. doi:10.1016/S0142-9612(02)00171-0
9. Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305–311. doi:10.1016/S1413-8670(11)70197-0
10. Růžička F, Hola V, Votava M, et al. Biofilm detection and the clinical significance of Staphylococcus epidermidis isolates. Folia Microbiol (Praha). 2004;49(5):596. doi:10.1007/BF02931540
11. Cookson BD, Robinson DA, Monk AB, et al. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J Clin Microbiol. 2007;45(6):1830–1837. doi:10.1128/JCM.02402-06
12. Donnio P-Y, Février F, Bifani P, et al. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob Agents Chemother. 2007;51(12):4342–4350. doi:10.1128/AAC.01414-06
13. Aires de Sousa M, Lencastre de H. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol. 2004;40(2):101–111. doi:10.1016/S0928-8244(03)00370-5
14. Carrico J, Pinto F, Simas C, et al. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 2005;43(11):5483–5490. doi:10.1128/JCM.43.11.5483-5490.2005
15. Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290(22):2976–2984. doi:10.1001/jama.290.22.2976
16. Oliveira DC, Milheiriço C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50(10):3457–3459. doi:10.1128/AAC.00629-06
17. Zhang K, McClure J-A, Elsayed S, Conly JM. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(2):531–540. doi:10.1128/AAC.01118-08
18. Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8(6):747–763. doi:10.1016/j.meegid.2008.07.007
19. David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. doi:10.1128/CMR.00081-09
20. Assafi MS, Mohammed RQ, Hussein NR. Nasal carriage rates of Staphylococcus aureus and CA-methicillin resistant Staphylococcus aureus among university students. Int J Microbiol Res. 2015;5(4):123–127.
21. Hussein NR, Basharat Z, Muhammed AH, Al-Dabbagh SA. Comparative evaluation of MRSA nasal colonization epidemiology in the urban and rural secondary school community of Kurdistan, Iraq. PLoS One. 2015;10(5):e0124920. doi:10.1371/journal.pone.0124920
22. Wayne P. Clinical and laboratory standards institute. Perform Stand Antimicrob Suscept Test. 2011.
23. Mirzaee M, Najar Peerayeh S, Ghasemian A-M. Detection of icaABCD genes and biofilm formation in clinical isolates of methicillin resistant Staphylococcus aureus. Iran J Pathol. 2014;9(4):257–262.
24. Mathur T, Singhal S, Khan S, Upadhyay D, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25. doi:10.4103/0255-0857.19890
25. Lotfi G, Hafida H, Nihel K, et al. Detection of biofilm formation of a collection of fifty strains of Staphylococcus aureus isolated in Algeria at the University Hospital of Tlemcen. Afr J Bacteriol Res. 2014;6(1):1–6. doi:10.5897/JBR2013.0122
26. Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–179. doi:10.1016/S0167-7012(00)00122-6
27. Zhang K, McClure J-A, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026–5033. doi:10.1128/JCM.43.10.5026-5033.2005
28. Omuse G, Van Zyl KN, Hoek K, et al. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: a cross sectional study. Ann Clin Microbiol Antimicrob. 2016;15(1):51. doi:10.1186/s12941-016-0171-z
29. Samadi R, Ghalavand Z, Nikmanesh B, et al. Investigation of biofilm formation among methicillin-resistant Staphylococcus aureus isolated from children. Arch Pediatr Infect Dis. 2018;6:3.
30. McClure J-A, Conly JM, Lau V, et al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant staphylococci. J Clin Microbiol. 2006;44(3):1141–1144. doi:10.1128/JCM.44.3.1141-1144.2006
31. Ghasemian A, Peerayeh SN, Bakhshi B, Mirzaee M. The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran J Pathol. 2015;10(4):258.
32. Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi:10.1086/313461
33. Dormanesh B, Siroosbakhat S, Darian EK, Afsharkhas L. Methicillin-resistant Staphylococcus aureus isolated from various types of hospital infections in pediatrics: panton-valentine leukocidin, staphylococcal chromosomal cassette mec SCCmec phenotypes and antibiotic resistance properties. Jundishapur J Microbiol. 2015;8:11. doi:10.5812/jjm
34. Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi:10.1086/596722
35. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016.
36. Gajdács M. The concept of an ideal antibiotic: implications for drug design. Molecules. 2019;24(5):892. doi:10.3390/molecules24050892
37. Singh SB, Young K, Silver LL. What is an “ideal” antibiotic? Discovery challenges and path forward. Biochem Pharmacol. 2017;133:63–73. doi:10.1016/j.bcp.2017.01.003
38. Kasim NA, Whitehouse M, Ramachandran C, et al. Molecular properties of WHO essential drugs and provisional biopharmaceutical classification. Mol Pharm. 2004;1(1):85–96. doi:10.1021/mp034006h
39. Yazdani R, Oshaghi M, Havayi A, et al. Detection of icaAD gene and biofilm formation in Staphylococcus aureus isolates from wound infections. Iran J Public Health. 2006;25–28.
40. Taj Y, Essa F, Aziz F, Kazmi SU. Study on biofilm-forming properties of clinical isolates of Staphylococcus aureus. J Infect Developing Countries. 2012;6(05):403–409. doi:10.3855/jidc.1743
41. Soto SM. Importance of biofilms in urinary tract infections: new therapeutic approaches. Adv Biol. 2014;2014.
42. Yousefi M, Pourmand MR, Fallah F, Hashemi A, Mashhadi R, Nazari-Alam A. Characterization of Staphylococcus aureus biofilm formation in urinary tract infection. Iran J Public Health. 2016;4(45):485.
43. Mohammed MK, Rasheed MN, Nadeer MI. detection of biofilm-associated genes in clinical Staphylococcus aureus isolates from Iraqi Patient. Int J Sci Nature. 2015;6(1):19–22.
44. Serray B, Oufrid S, Hannaoui I, et al. Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. J Infect Developing Countries. 2016;10(08):863–869. doi:10.3855/jidc.8361
45. Takano T, Higuchi W, Otsuka T, et al. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob Agents Chemother. 2008;52(3):837–845. doi:10.1128/AAC.01001-07
46. Eady EA, Cove JH. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus– an emerging problem for the management of skin and soft tissue infections. Curr Opin Infect Dis. 2003;16(2):103–124. doi:10.1097/00001432-200304000-00007
47. Moghadam SO, Pourmand MR, Douraghi M, Sabzi S, Ghaffari P. Utilization of PFGE as a powerful discriminative tool for the investigation of genetic diversity among MRSA strains. Iran J Public Health. 2017;46(3):351.
48. Hussein NR. Prevalent genotypes of Staphylococcus aureus strains isolated from healthcare workers in Duhok City, Kurdistan Region, Iraq. Int J Infect. 2016;3:2. doi:10.5812/iji.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
