Back to Journals » Drug Design, Development and Therapy » Volume 17
Antimalarial Drug Resistance: A Brief History of Its Spread in Indonesia
Authors Fitri LE , Pawestri AR , Winaris N , Endharti AT , Khotimah ARH , Abidah HY , Huwae JTR
Received 20 January 2023
Accepted for publication 25 April 2023
Published 5 July 2023 Volume 2023:17 Pages 1995—2010
DOI https://doi.org/10.2147/DDDT.S403672
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Manfred Ogris
Loeki Enggar Fitri,1,2 Aulia Rahmi Pawestri,1,2 Nuning Winaris,1,2 Agustina Tri Endharti,1 Alif Raudhah Husnul Khotimah,3,4 Hafshah Yasmina Abidah,3,4 John Thomas Rayhan Huwae3,5
1Department of Parasitology Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia; 2AIDS, Toxoplasma, Opportunistic Disease and Malaria Research Group, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia; 3Master Program in Biomedical Science, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia; 4Medical Doctor Profession Education, Faculty of Medical and Health Science, Maulana Malik Ibrahim State Islamic University, Malang, Indonesia; 5Medical Doctor Profession Study Program Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia
Correspondence: Alif Raudhah Husnul Khotimah, Master Program in Biomedical Science, Faculty of Medicine, Universitas Brawijaya, 65145, District Lowokwaru, Malang, East Java, Indonesia, Tel +62 822 573 458 27, Fax +62 341 564755, Email [email protected]; [email protected]
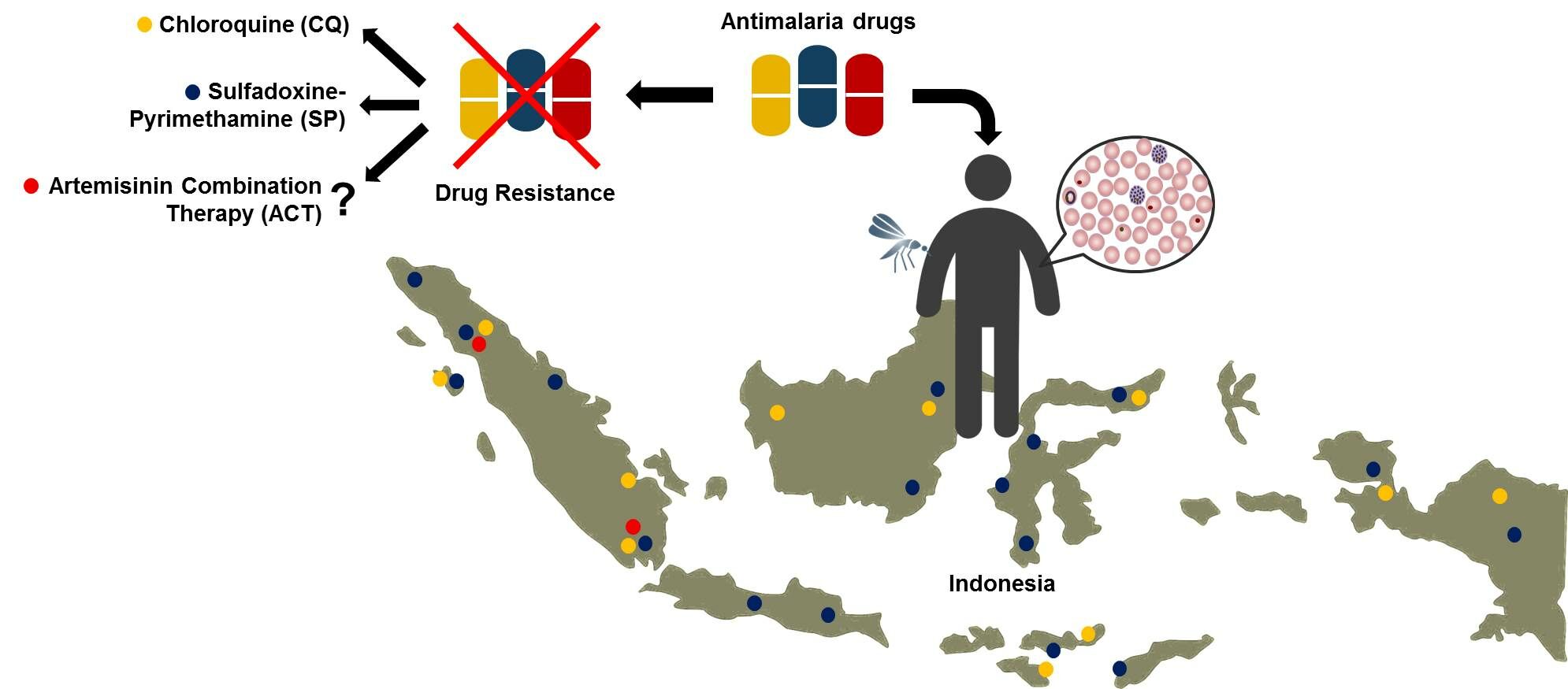
Abstract: Malaria remains to be a national and global challenge and priority, as stated in the strategic plan of the Indonesian Ministry of Health and Sustainable Development Goals. In Indonesia, it is targeted that malaria elimination can be achieved by 2030. Unfortunately, the development and spread of antimalarial resistance inflicts a significant risk to the national malaria control programs which can lead to increased malaria morbidity and mortality. In Indonesia, resistance to widely used antimalarial drugs has been reported in two human species, Plasmodium falciparum and Plasmodium vivax. With the exception of artemisinin, resistance has surfaced towards all classes of antimalarial drugs. Initially, chloroquine, sulfadoxine-pyrimethamine, and primaquine were the most widely used antimalarial drugs. Regrettably, improper use has supported the robust spread of their resistance. Chloroquine resistance was first reported in 1974, while sulfadoxine-pyrimethamine emerged in 1979. Twenty years later, most provinces had declared treatment failures of both drugs. Molecular epidemiology suggested that variations in pfmdr1 and pfcrt genes were associated with chloroquine resistance, while dhfr and dhps genes were correlated with sulfadoxine-pyrimethamine resistance. Additionally, G453W, V454C and E455K of pfk13 genes appeared to be early warning sign to artemisinin resistance. Here, we reported mechanisms of antimalarial drugs and their development of resistance. This insight could provide awareness toward designing future treatment guidelines and control programs in Indonesia.
Keywords: Indonesia, antimalarial resistance, chloroquine, sulfadoxine-pyrimethamine, primaquine, artemisinin
Graphical Abstract:
Introduction
The history of malaria in Indonesia began in the 1900s when the number of patients reached 30 million and causing mortalities in 120,000 people.1–3 Accounting 15.6% of cases and 22% of malarial deaths, Indonesia has been declared as one of the malaria endemic countries in South-East Asia.4
The most common Plasmodium species in Indonesia is Plasmodium falciparum (P. falciparum), followed by Plasmodium vivax (P. vivax). Both species occasionally co-distribute sympatrically in several areas of Indonesia. A lesser common species, Plasmodium malariae (P. malariae), is found on all the main islands, with the eastern part of Indonesia reporting the majority of cases. Meanwhile, reports of Plasmodium ovale (P. ovale) mostly stemmed exclusively from the eastern part of Indonesia.5 In 2022, according to the Directorate of Prevention and Control of Infectious Diseases, Ministry of Health Indonesia, there were 399,666 confirmed cases of malaria in Indonesia, with 51.3% of them being caused by P. falciparum, 33.4% by P. vivax, and the remaining by other Plasmodium species.6
In Indonesia, chloroquine-resistant cases of malaria were first reported in East Kalimantan in early 1974.7 Since the 1990s, cases of resistance have spread to all provinces. In addition, there have also been reports of resistance to sulfadoxine-pyrimethamine (SP) in several areas.5,8 Artemisinin resistance has never been reported in Indonesia, although recently the spatial distribution of its resistance marker, the K13 propeller polymorphism, has been reported in Asia, as validated in India in 2019, and in the Greater Mekong subregion since 2006.9
Numerous antimalarial drug classes are currently on the market; however, due to toxicity concerns, reduced efficacy, and, most importantly, resistance, these drugs have decreased their overall therapeutic indices.10 By performing literature search of papers published in public databases using relevant keywords, this review laid over some impetus, particularly focusing on the growing antimalarial resistance of P. falciparum and P. vivax. We also discussed the mechanism of malaria drug resistance in general and the development and spread of anti-malarial resistance in Indonesia.
Malaria in Indonesia
The detailed history of malaria program evolution in Indonesia has shown particularly positive progress in the last decades, with over 50% of the districts formally deemed malaria-free in 2017. Between 2007 and 2017, the annual parasite incidence (API) decreased from 2.89 to 0.9 per 1000 population, accounting for over threefold. This decline was consistent with a 50% and 66% reduction in confirmed malaria cases and mortalities, respectively.11 Recently, WHO estimated the number of people with malaria infections decreased from 1.7 million in 2008 to 1 million in 2018. This decline was also consistent with the shrinkage of areas with malaria transmission.12 According to the recent WHO report, as the fourth most populated country, Indonesia had an estimate of 800,000 malaria cases in 2021, which ranked second in Asia after India.13
In 2010, there were a total of 6,699 districts in Indonesia, with 450 districts suspected of having malaria cases; however, 266 have successfully become malaria-free, 93 have downgraded from high to moderate or low transmission, and 38 had at least threefold reduction in API.11,12 Currently, over 55% of districts in Indonesia are free from local transmission and 72% of the population lives in malaria-free areas.11,12 According to the National Malaria Program Report 2018, 91% of malaria cases are reported from only three provinces: East Nusa Tenggara, Papua, and West Papua. From January to March 2020, East Nusa Tenggara, Papua, and West Papua reported 266,956 suspected cases of malaria which were tested by microscopy/rapid diagnostic test (RDT), yielding a total of 23,632 positive cases (8.9%). These results were reduced when compared to the same period in the previous year, when 677,536 suspected cases were tested and 64,753 positive cases were confirmed (9.5%).12
Since 2004, Indonesia has proclaimed the first-line anti-malarial drug regimen using artemisinin-based combination therapy (ACT) to replace the ineffective chloroquine and sulfadoxine–pyrimethamine (SP). Nevertheless, the expeditious appearance and spread of resistant Plasmodium strains to commonly used anti-malarial drugs, such as chloroquine, sulfadoxine–pyrimethamine, and artemisinin-based combination therapy, continue to pose a threat to the national malaria elimination programs.14
Antimalarial Drug Resistance Mechanisms
Antimalarial resistance is defined as the ability of a parasite strain to live and/or proliferate even when a drug is administered and absorbed at dosages that are higher or equivalent to those typically advised but within the subject’s tolerance.15 A change in a protein’s composition, structure, or function appears to be the underlying cause of resistance.16 To detect the establishment of resistance and gauge its spread, antimalarial drug resistance molecular markers are utilized. It details the genetics of parasites linked to resistance, such as gene copy number variations or single nucleotide polymorphisms, that lead to a reduction in parasite sensitivity to antimalarial medications.17 Table 1 shows the mechanisms of all antimalarial drugs and resistance markers. Major antimalarial drugs and their mechanisms of resistance are illustrated in Figure 1.
 |
Table 1 Antimalarial Drug Mechanism and Resistance Marker |
4-Aminoquinoline (Chloroquine and Piperaquine)
The asexual malaria parasite grows and develops in host erythrocytes by digesting hemoglobin in its acidic food vacuoles, a process that produces amino acids, free radicals, and heme (ferriprotoporphyrin IX/FPIX). FPIX is toxic to parasites, so it is converted into a polymer hemozoin or known as malaria pigment. The chloroquine base diffuses into the food vacuole and becomes protonated and trapped within the vacuole. Chloroquine binds tightly to FPIX to form the toxic FPIX-Chloroquine complex that inhibits the formation of hemozoin, poisons the acidic food vacuoles and starves the parasite. The accumulation of chloroquine also increases the pH in the vacuole, which reduces the polymerization rate of FPIX.18
In Indonesia, chloroquine was used as a first-line treatment in the national malaria elimination program in 1959 after the Indonesian independence. It was used until 2004 before being replaced by ACT.19 Resistance of P. falciparum to chloroquine and possibly amodiaquine are associated with mutations in pfcrt that encode the P. falciparum chloroquine resistance transporter (PfCRT).20,21 Mutations in pfcrt permit transporters to move chloroquine out of the digestive vacuole and away from the drug’s anti-plasma targets (heme detoxification).22 The primary mechanism of this resistance is mediated by an energy-dependent drug efflux mechanism mediated by PfCRT, resulting in decreased chloroquine accumulation in digestive vacuole.18,21,23 A single nucleotide polymorphism (SNP), K76T, in pfcrt is correlated with in vitro chloroquine resistance. Reversal of the K76Tmutation resulted in wild-type CQ susceptibility and led to increased binding of CQ to FPIX. In the presence of chloroquine compared to sensitive parasites, parasites carrying the K76T mutation exhibit an advanced leak of H+ from the digestive vacuole. Other polymorphisms of pfcrt, including 184F, 1034C, 1042D, and 1246Y, are also associated with chloroquine resistance to varying degrees.24
Another chloroquine resistance mechanism is correlated with P. falciparum multidrug resistance transporter 1 (PfMDR1). PfMDR1, found in the digestive vacuole, functions as a general transporter and binds toxic metabolites or drugs to the digestive vacuole.17 This pump imported antimalarial drugs and uptakes solutes into the digestive vacuole.17,20,24 Mutation of pfmdr1 N86Y is associated with decreased chloroquine sensitivity in vitro. However, the pfmdr1 mutation alone is not sufficient to cause chloroquine resistance.20
Piperaquine inhibits one or more steps in hemoglobin breakdown, thus free and undigested hemoglobin are accumulated and the formation of hemozoin is inhibited.25 Resistance to piperaquine is correlated with gene plasmepsins pfpm2 and pfpm3.21,26 It was proposed that overproduction of these plasmepsins might interfere with piperaquine’s inhibition of heme detoxification processes in the digestive vacuole.21,26 However, it could not be confirmed whether plasmepsin is directly involved in mediating piperaquine resistance or was a compensatory mechanism for the fitness disadvantage of the actual resistance determinant.21
Plasmodium falciparum piperaquine resistance is influenced by multiple factors, mainly the polymorphisms of pfcrt and pfmdr1. The genetic mediator of piperaquine resistance in P. falciparum is the SNP of pfcrt (C101F, H97Y, F145I, M343L, G353V), plasmepsin 2 and 3 amplification, and pfmdr1 single copy.20 Piperaquine-resistant parasites have a reduced ability to inhibit hemoglobin digestion through the amplification of plasmepsin 2 and/or plasmepsin 3, leading to reduced drug accumulation within digestive vacuole, primarily mediated by polymorphisms in pfcrt and pfmdr1.25 In Indonesia, piperaquine is used in combination with dihydroartemisinin (dihydroartemisinin-piperaquine) as the first-line treatment for uncomplicated falciparum malaria since 2010.27 Resistance to dihydroartemisinin-piperaquine is also associated with P. falciparum exonuclease (Pfexo) and pfcrt gene polymorphisms. Resistance to both dihydroartemisinin and piperaquine found in the Southeast Asian parasites exhibit simultaneous polymorphism in P. falciparum Kelch 13 (PfK13) and amplification of the parasitic proteases plasmepsin 2 and 3.18
8-Aminoquinoline (Primaquine and Tafenoquine)
Primaquine and tafenoquine are the 8-aminoquinoline drugs used to treat and prevent malaria relapse. Based on the Indonesian Ministry of Health guideline on malaria elimination, primaquine in combination with dihydroartemisinin-piperaquine is still used in Indonesia, particularly for malaria caused by P. vivax or P. ovale. Tafenoquine single dose has been approved to prevent relapses of P.vivax.20 Tafenoquine's mechanism of action is still completely unclear, and there are currently no recognized molecular targets for this or other 8-aminoquinolines. Unlike primaquine, tafenoquine possesses a 3-(trifluoromethyl)phenoxy group that inhibits position 5 from being directly oxidized, resulting in delayed elimination and long-acting action.114 Primaquine can inhibit the stage V gametocytes of P. falciparum and thus effectively decrease malaria transmission.20 Primaquine also prevent recurrent infections of P. vivax by targeting the latent stage of the parasite in the liver.17 Identifying primaquine resistance in P. vivax is challenging due to possibilities of reinfection in malaria-endemic areas.20
A variety of human and parasite variables are thought to be involved in modulating primaquine’s ability to kill hypnozoites, including treatment adherence and human genetic polymorphisms.28 Primaquine is a prodrug that must be metabolized to produce active molecules via the action of CYP2D6 and CPR.29 Studies have shown that various common polymorphisms of these enzymes can increase or reduce enzymatic activity.30 Polymorphisms in CYP2D6 that decrease enzyme activity may result in impaired primaquine metabolism and treatment failure.31 Similar to the P. falciparum resistance against artemisinin, hypnozoites ability to resist oxidative stress would support primaquine resistance and increase difficulties in P. vivax eradication.32,33
Naphthoquinone
Atovaquone, a naphthoquinone drug class, is commonly used as a combination therapy with an antifolate drug such as atovaquone-proguanil (Malarone). Although not used as a first-line treatment in Indonesia, it has been well reported in some Indonesian populations.34 The target of atovaquone is a mitochondrial electron donor, P. falciparum cytochrome b (PfCytB).18 PfCytB donates electrons to dihydroorotate dehydrogenase (DHODH), an enzyme involved in de novo pyrimidine synthesis that is critical for blood-stage asexual parasites.20 Resistance to atovaquone monotherapy evolves rapidly and single-point mutations in the genes encoding cytochrome b in Plasmodium sp.21,35 pfcytb gene, such as Y268S/C/N, leads to alterations in its catalytic activity and is linked to clinical failure of atovaquone. Although atovaquone resistance may emerge, pfcytb mutant parasites exhibit a reduced transmission to mosquito, so that atovaquone resistance might not spread quickly.18,20,21
Antifolate
Antifolate antimalarial drugs including proguanil, pyrimethamine, trimethoprim sulfadoxine, and sulfamethoxazole.17,20,35 In Indonesia, antifolates are currently used as combination therapies such as atovaquone-proguanil for prophylaxis, particularly in pregnant women or children, or sulfadoxine-pyrimethamine to treat P. falciparum.20,36 Sulfadoxine is a structural analog of para-amino benzoic acid (pABA) and can inhibit dihydropteroate synthase (DHPS), while pyrimethamine inhibits dihydrofolate reductase (DHFR), essential enzymes for folate synthesis for the malaria parasite.37 Sulfadoxine-pyrimethamine also has antimitotic effects on erythrocytes and tissues, and its action is prolonged by sulfadoxine.36
Reduced efficacy of sulfadoxine-pyrimethamine during P. falciparum treatment is associated with increased prevalence of mutations in the antifolate resistance genes dhps and dhfr,35,36 which causes reduced drug-binding affinity of DHFR.24,36 These are amino acid substitutions at codons S108N, N51I, C59R, and I164L25, and additional point mutations in I51, R59, and L164.24 Sulfadoxine resistance is related to variations in the dhps gene at codons S436A/F, A437G, K540E, A581G, and A613T/S.36
Amino Alcohol
Mefloquine and lumefantrine, quinine derivatives from the aryl amino alcohol group, are currently used in conjunction with artemisinin derivatives. Mefloquine is also prescribed by itself as a prophylaxis.20 Quinine binds reactive heme and prevents it from being detoxified by integrating it into chemically inert hemozoin, which occurs in the digestive vacuole.38 Quinine resistance is controlled by PfCRT and PfMDR1, which possibly comprise an enzyme that participates in the ubiquitination of protein.39 P. falciparum’s susceptibility to quinine often changes in its sensitivity to chloroquine, amodiaquine, or mefloquine in unexpected ways, such as hypersensitivity or causing cross-resistance.40 These findings indicated the mechanism of quinine in the partial suppression of heme detoxification.38
An increase in the number of copies of pfmdr1 was found to be the main factor mediating resistance to mefloquine.41–43 The pfmdr1 copy number variation (CNV), or considerable tandem amplification up to 100 kb, encompasses a large number of genes. It is notable that amplicon break sites are often located in intergenic regions of monomeric tracts A or T in P. falciparum CNVs. Given the high AT content and many AT monomeric tracts in the genome of P. falciparum, CNVs are a crucial route of evolutionary adaptation.20
Antibiotics
Although less common in Indonesia, antibiotics including azithromycin,44 clindamycin,45 and doxycycline46,47 have been used to treat and prevent malaria, by preventing the ribosomes from functioning normally in the apicoplast and mitochondria of Plasmodium sp., resulting in lowered DNA gyrase activity or protein synthesis.48,49 The defective apicoplasts are passed to their offspring, which had a potent but delayed antimalarial impact. Based on the existence of potential apicoplast targeting signals, hundreds of nuclear-encoded proteins are projected to localize there.18,49
Azithromycin and clindamycin have potent antimalarial activity via an action against the apicoplast organelle.45,50 However, doxycycline instead caused non-functional apicoplasts to disperse throughout the progeny of treated parasites after treatment, preventing them from completing cell division.49 Apicoplast ribosomal RNA alterations mediate P. falciparum resistance.20,38 The mechanism behind clindamycin resistance has been identified as a point mutation encoded 23S rRNA in the apicoplast, which exhibits a “delayed death” phenotype.17,51
Sesquiterpene Lactone Endoperoxides
All P. falciparum asexual stages are swiftly eliminated by artemisinin, the current most effective antimalarial drug.52 The effectiveness and early action of artemisinin and its derivatives, such as artemether, artesunate, dihydroartemisinin (DHA), and artemether, as well as synthetic artemisinin compounds, cause the parasitemia to rapidly decline within the first few days of treatment. As a part of ACTs, these drugs are currently used as the first-line treatment in Indonesia and worldwide.17,20 These chemicals are activated by parasites when their endoperoxide bridge is broken, which results in oxidative stress. When artemisinin is active in the asexual blood stages of parasites, Fe2+-heme reductively scissions their endoperoxide bridge.53 A host red cell-derived enzyme called superoxide dismutase catalyzes the fast conversion of heme-FeII (LH) (released from degraded Hb within the food vacuole) to heme-FeIII (hematin/hemin) and superoxide anion (O2−), the latter of which disproportions into H2O2 and O2 (ingested from red cell cytosol).54 This results in ACTs killing parasites when they are in an active condition.53
The primary contributor to artemisinin resistance has been identified as the pfk13 gene on chromosome 13.55–57 It is believed that the K13 protein contributes to the cellular reaction to oxidative stress.53 It is unclear what particular functional alterations the K13 polymorphisms cause; however, resistant parasites have a better cell stress response during the early ring stages when artemisinin is most active. They also exhibit an increased stress response, lower levels of ubiquitinated proteins, and a delayed start to cell death.58 According to research, the K13 C580Y variation reduces the interactions between Plasmodium falciparum phosphatidylinositol-3-kinase (PfPI3K) and artemisinin, which lowers polyubiquitination by PfPI3K and lowers phosphatidylinositol-3-phosphate (PI3P), a component of phospholipid signaling.59 According to African research, K13 levels were low in highly resistant variants and high in low resistant mutations. It reveals that various occurrences favoured creating different K13 changes, explaining the mutations congregated in two groups, one with high resistance-low K13 levels, which includes the C580Y mutation, and another with low resistance-high K13 levels, which includes the V520A and V589I mutations.60
Detection of Antimalarial Drug Resistance
Antimalarial drug resistance has been a persistent hindrance for malaria treatment for years, not only in Indonesia, but also worldwide. Evaluation for drug resistance is highly recommended to determine the success of malaria treatment.61 The identification of antimalarial drug resistance in Plasmodium species can be performed through several methods, including in vivo, drug level, in vitro, ex vivo, and molecular characterization.62,63
In 2009, WHO established a protocol to detect antimalarial drug resistance.62 The protocol incorporates recommendations from all malaria endemic areas as well as the use of clinical and microbiological studies to evaluate the success of treatment and detection of drug resistance.64 The same protocol is currently being used in Africa as stated by the WHO guideline 2022 to overcome antimalarial drug resistance.65 The first method described in the WHO protocol is an in vivo test. This method is designed to clinically evaluate the efficacy of antimalarial drugs. Throughout in vivo study, patients with malaria are prescribed the first-line antimalarial drugs with standard dose, and the parasitemia levels are monitored through both clinical and laboratory assessments to evaluate the response to therapy.51 This method is widely-known as therapeutic efficacy studies (TES) which is important to investigate the mechanism of drug resistance, where the host–parasite interactions is suggested to contribute in altering the susceptibility of the antimalarial drug.63 TES is also useful to measure the drug efficacy in real time, which proved to be beneficial particularly in multispecies or recurrent infection cases. The outcomes of TES are evaluated at the end of the study, usually on day 28 or 42 (depending on the half-life of the drugs). Furthermore, genotype examination is required to perform when the infections occurred during the follow-up period, thus enabling us to distinguish between the new and recurrent infections.65
The second method described in the WHO guideline is a test to measure the level of antimalarial drug collected from the blood of the malaria patients who consumed antimalarial drug. The concentration of the drug, which is extracted from the blood, is analyzed using high-performance liquid chromatographic (HPLC) method. Evaluation on the drug level in the blood can help to differentiate between medication nonadherence and drug resistance.66
The third method described by WHO is in vitro and ex vivo tests, which eliminate the host–parasite interactions and simply observe the efficacy of antimalarial drugs on the Plasmodium itself.63 In principle, these methods use Plasmodium parasites obtained from malaria patients, which are cultured and exposed to the different antimalarial drugs with different concentrations. The growth and survival of Plasmodium are evaluated to determine the drug susceptibility and resistance profile.64 There are several approaches used to measure the parasite’s growth and survival rate in in vitro and ex vivo tests. For example, a simple Giemsa staining is easily performed on the culture smear and the number of surviving parasites is manually counted under the microscope to determine the half maximal inhibitory concentration (IC50). However, this method is considered to be impractical, as it is time and labor consuming in clinical settings where numerous samples need to be examined daily.62 Other alternative approaches are also available, including: 1) isotopic tests using radioactive dyes, 2) flow cytometry to evaluate the number of infected erythrocytes, and 3) enzyme-linked immunosorbent assay (ELISA) to quantify the concentrations of proteins, such as histidine rich protein II (HRP-2), which is produced by the Plasmodium sp. and accumulated in the culture.63
Another method that can detect antimalarial drug resistance is molecular characterization, which has progressively developed in recent years.67 Molecular characterization is used to identify the markers of antimalarial drug resistance. It can be divided into two different methods, which are polymerase chain reactions (PCR) and sequencing. PCR-based methods are mainly used to identify short amplicons of antimalarial drug resistance genes in the laboratory. These PCR methodsinclude restriction fragment length polymorphism (RFLP), sequence-specific probe-based ELISA, ligase detection reaction fluorescent microsphere (LDR-FM), and real time-PCR. Moreover, sequencing methods are performed using either Sanger or next-generation sequencing (NGS), such as Illumina MiSeq, Pyrosequencing, and MinION nanopore, which are suitable to identify numerous genetic markers for antimalarial drug resistance in a large number of samples for each run.61,67–69
Application of various detection methods in antimalarial drug resistance depends on the facility in malaria endemic areas. In Indonesia, TES is commonly used for the studies of drug resistant,8,70,71 as well as molecular methods, such as PCR (nested and RFLP).72–74 However, there were no specific regulations issued by Indonesian government that provide a standard procedure or guideline for detection of antimalarial drug resistance. Although each method has its own advantages and disadvantages, the different uses and applications allow the vast development in antimalarial drug resistance studies.63 Furthermore, a comparison study among different methods used to detect antimalarial drug resistance may be advantageous for the management of malaria treatment and epidemiological study in the future.
Antimalarial Drug Resistance in Indonesia
Resistance to 4-Aminoquinoline (Chloroquine)
After its discovery in the 1930s, chloroquine became widely used as a first-line monotherapy for malaria in Indonesia. In 1957, the first case of chloroquine resistance was reported in Thailand,75 which eventually spread to other regions of the world. Many cases of chloroquine resistance in Indonesia were imported and occurred due to the influx of travelers from the Southeast Asian region to Indonesia.75–77 Despite the growing concerns and evidence of resistance and the introduction of ACT, chloroquine was continuously used until the Indonesia Ministry for Health removed its recommendation for chloroquine monotherapy in 2013.8 This part discusses the timeline of chloroquine resistance in Indonesia during the span of 1974–2015 (shown in Figure 2).
In Indonesia, the first documented case of chloroquine-resistant P. falciparum occurred in Kalimantan in 1974.8 This case involved immigrants from Japan. In 1974 a case of P. falciparum recrudescence was also reported in West Irian (now known as Papua), which also involved migrants from Japan. Resistance was determined based on clinical symptoms, plasma chloroquine levels, and in vivo and in vitro sensitivity tests. At the same time, several cases of resistance from local immigrants were reported in East Kalimantan based on clinical symptoms and in vivo sensitivity tests.7,78,79 In the early 1990s, more cases were further reported in East Kalimantan by in vitro test.80,81
During the span of 1992 and 1994, chloroquine-resistant P. vivax was observed in the Javanese population in Papua who showed 44% and 78% of chloroquine treatment failure within 14 and 28 days of initiating therapy. This population, however, still showed sensitivity toward the combination of chloroquine plus primaquine or halofantrine alone.82 In 1995, a survey in the central northern coasts of Papua revealed that, despite adequate levels of chloroquine’s active metabolite in the circulation, the cumulative treatment failure rates reached of 45% and 58% at 14-day post initiation of therapy, which increased to 64% and 89% at day 28, respectively.83 Similarly, in the same year, a study in north-eastern Papua found 95% of P. falciparum, 84% of P. vivax, and 100% of mixed infections, failed to respond to chloroquine therapy.84 Furthermore, West Kalimantan reported cases of P. vivax infections that were resistant to chloroquine three years later, where in vivo studies found recurring parasitemia during the 28-day observation period.85
During 2001 and 2002, a study in Alor, Nusa Tenggara, showed a cumulative incidence of chloroquine resistance of 56% at day 28 post-treatment initiation, and thus ending the use of this drug as antimalarial in Alor.86 At the same period, cases of failure of chloroquine treatment against P. vivax and P. falciparum were recorded in Lampung, Sumatra, with a 28-day cumulative incidence of 43% and 68%, respectively.87
A study testing samples collected in 2003 in Timika, Papua, showed that pvmdr1 gene polymorphism with a Y976F mutation was found in 96% of samples, which displayed 65% chloroquine resistance using the modified schizont maturation assay. This study thus suggested the Y976F variation of pvmdr1 as a predictive marker for chloroquine resistance.88
In 2004, early chloroquine treatment failure of P. vivax and P. falciparum cases was reported at 16% and 5%, respectively, in Timika, Papua. The failure rate of P. vivax treatment after 28 days reached 73%, while the 42-day failure in P. falciparum was 48%, despite it still being effective for P. ovale and P. malariae.89
In 2007, 25% of studied patients in the north-eastern part of Papua showed persistent or recurrent parasitemia of P. vivax following chloroquine treatment, with a 17.5% probability of parasite resistance.72 In the same year, seasonal distribution of gene polymorphisms linked to chloroquine resistance against P. falciparum was found in West Sumba. Mutant pfmdr1 alleles, predominantly as 1042D and 86Y, were found higher in the wet seasons.90
A study using samples collected from 2010 to 2011 from many areas of Sulawesi and Kalimantan was carried out to identify the P. vivax mutations related to chloroquine-resistance. They discovered that 93% of samples harbored the Y976F variation of the pvmdr1 gene, which was correlated with a reduced chloroquine sensitivity in Papua.91 In the same year, a study in North Sulawesi found polymorphisms of the chloroquine-resistance genes in P. falciparum, pfcrt 76T, in 76.4% of tested isolates, while pfmdr1 86Y and 86N were observed in 88.2% and 10.2% isolates, respectively.74 Moreover, in 2012, a study in South Sumatera identified polymorphisms of the pfcrt 76T and pfmdr1 86Y in all of their tested isolates.92
Finally, in 2015, a large-scale survey involving 404 molecularly confirmed P. falciparum infections found that the pfcrt haplotype SVMNT (codons 72 to 76) showed the most prevalent and pronounced linkage disequilibrium with the pfmdr1 haplotype YY (codons 86 and 184) although the parasites’ persistence and increased at day 28 or 42 post-treatment initiation were significantly more likely to harbor the pfmdr1 haplotype NF (codons 86 and 184).93 There have been no further reports after nearly a decade of understanding that the use of chloroquine as monotherapy was inappropriate.
As in Indonesia, in Southeast Asia using pfcrt and pfmdr1 mutation measurement and gene copy number has been found to be useful for surveillance of mefloquine-resistant malaria.94 One Southeast Asian nation, Cambodia, has seen the emergence of novel pfcrt mutations that cause resistance to dihydroartemisinin plus piperaquine (DHA + PPQ).95 In Southeast Asia, the pfmdr1 gene, which encodes the N86 and 184F haplotypes, is frequently found to have multiple copies. Besides that, parasites that express this PfMDR1 variant are known to respond differently to antimalarial medications due to their increased transport capacity.96
Resistance to Antifolate
Although not being used as first-line antimalarial treatment, the combination of sulfadoxine and pyrimethamine is widely used both prophylactically and therapeutically, particularly in chloroquine-resistant areas. Thus, it was not surprising when in 1979, the first occurrence of sulfadoxine-pyrimethamine resistance was reported in Indonesia.70 As with other antimalarial resistance, this further spread over the next few years to other areas in Indonesia, where four provinces (Central Java, East Timor, South Sulawesi, and Papua) reported resistance of sulfadoxine-pyrimethamine to P. falciparum in the span of 1981–1995, as proven by in vivo assays (Figure 3).8
A study was conducted in 1996–99 to observe the relationship between mutations in the P. falciparum pfmdr1, dhfr and dhps genes and in-vivo drug resistance in West Papua. Initially, 85 patients infected with P. falciparum were treated with chloroquine, of which 21 had clearance of the parasites, 49 had parasitemia classified as RI (clearance of asexual parasite in 7 days followed by recrudescence within 28 days), RII (more than 75% clearance of asexual parasitemia within 48 hours), or RIII (less than 75% reduction in asexual parasitemia within 48 hours) resistance, while 15 patients were excluded from statistical analysis because of incomplete clinical histories. Sulfadoxine-pyrimethamine was then used for the second-line drug, which resulted in parasite clearance in 18 patients, while 31 had continuing infections classified as RI, RII, or RIII resistance. pfmdr1 mutants are associated with chloroquine resistance and dhfr and dhps mutants are associated with in vivo sulfadoxine-pyrimethamine resistance. It is interesting to note that 437G in dhps and Arg-59/Asn-108 in dhfr were linked to RI, RII, and RIII resistance, while 540Q was only strongly related to RII and RIII resistance. This result supports the theory that the molecular basis of RI, RII, and RIII sulfadoxine pyrimethamine resistance is the accumulation of dhfr and dhps mutations.97
By using samples collected in Papua in 1996–1999 and Central Java in 2000, a study evaluating the effect of P. vivax dhfr polymorphism toward sulfadoxine-pyrimethamine resistance reported that 58% of cases had poor response to treatment. Across 6 identified alleles, they found 2 novel mutations, including 99S and 19V. One allele carrying four point mutations (57L+58R+61M+117T) also increased the risk of developing early treatment failure to 23-fold.98 Meanwhile, a survey conducted in 8 endemic areas in Indonesia identified that polymorphisms in four genes were related to resistance in P. falciparum, including dhfr (16V, 59R, and 108N/T), dhps (437G or with 540E), pfcrt (76T), and pfmdr1 (86Y, 1042D).99
A 28-day in-vivo test was carried out on 167 villagers from the Menoreh Hills of Central Java. Chloroquine and sulfadoxine-pyrimethamine were studied as first- and alternate-line drug for non-complicated malaria in Indonesia. Malaria was found in 33% of the 1389 residents who were screened prior to enrollment. Microscopic diagnosis, PCR-based confirmation, concentrations of chloroquine and desethylchloroquine in blood, and P. falciparum genotype identification were used to evaluate treatment outcomes. For P. falciparum treatment, the 28-day cumulative incidences of therapeutic failure for chloroquine and sulfadoxine-pyrimethamine were 47% and 22%, respectively, and for P. vivax treatment, they were 18% and 67%.100
Another study in Nias Island, North Sumatra found that active case detection revealed malaria cases in 124 (17%) of 710 local residents, while passive case detection identified malaria cases in 77 (44%) of 173 suspected clinical malaria patients. P. falciparum was treated with sulfadoxine-pyrimethamine on day 0, whereas P. vivax patients were treated with chloroquine on days 0, 1, and 2. The clinical and parasite responses were tracked until day 28. Recurrent parasitemia was observed in 29 (83%) of the 35 P. falciparum cases treated by day 28, with four cases showing RI, RII, and RIII resistance. Six (21%) of the 28 P. vivax cases between days 11 and 21 showed recurrent parasitemia. Mutations associated with P. falciparum infections were discovered when the parasites present in patients with P. falciparum infections were genotyped.101
During a two-stage survey conducted in 2004–2006 and 2009–2012 in Kalimantan and Sumbawa, West Nusa Tenggara, polymorphisms in the pfdhfr (C59R, S108N, and I164L) and pfdhps gene (A437G, K540T, A581G, and I588F) were identified. The pfdhps K540T was hypothesized to originate in Kalimantan and I558F in Sumbawa, and played a role in the transmission of sulfadoxine-pyrimethamine-resistant P. falciparum in other areas in Indonesia.102
In 2006, early sulfadoxine-pyrimethamine and chloroquine treatment failure was also observed in 4% of P. falciparum- and 15% P. vivax-infected patients in Southern Papua. Further recurrence was found in 48% and 70% of P. falciparum and P. vivax infections, respectively.71 In 2006–2008, a survey was conducted to map the distribution of alleles related to sulfadoxine-pyrimethamine-resistant P. vivax in Papua, Central Java, and Sumba, East Nusa Tenggara. The dhps 383G was detected in allele frequencies ranging from 9% to 33% in these regions. The dhfr 57L/I, 61M, and 117N/T polymorphisms were also found in Lampung, Central Java, Sumba, and Papua.103
Between 2008 and 2012, a study in P. falciparum-infected pregnant women in South Kalimantan found important mutations in the pfdhfr (N51, S108N) and pfdhps genes (A437G, K540 and A581), where pfdhfr 108N and pfdhps 437G were found in all pregnant women with P. falciparum infection.73 Similarly, during 2009–2010, pfdhps K540T and I588F mutations were found in 61 P. falciparum patients in South Kalimantan, indicating their importance in the development of sulfadoxine-pyrimethamine resistance.102
In Southeast Asia, except for Indonesia, publications that particularly address sulfadoxine-pyrimethamine resistance caused by the pfdhfr and pfdhps gene mutation are quite scarce. A polymorphism of I164L was reported to increase resistance to pyrimethamine to a point where sulfadoxine-pyrimethamine might no longer be effective. However, only one sample from Myanmar had been identified to harbor the I164L mutation in 2011 (0.3%, 1/300).104,105
Resistance to Sesquiterpene Lactone Endoperoxides
Due to the growing resistance to monotherapy antimalarial drugs, in 2006, the malarial treatment guideline in Indonesia had been revised to use ACT as a first-line therapy for uncomplicated malaria. Despite its good efficacy in promoting a trend of malaria control, several cases of treatment failures have been suspected in Indonesia (Figure 4).
In 2009, persistent parasitemia in 2 out of 103 (1.9%) P. falciparum-infected patients from West Sumba showed persistent parasitemia until day 7 and increased parasitemia at day 28 post-treatment with artesunate-amodiaquine. Both patients showed different glutamate-rich protein(GLURP) genotypes on days 21 and 28, which indicated re-infection with a new strain of P. falciparum, suggesting that artesunate-amodiaquine was still effective in this area.106
In 2015, genotyping of the pfk13 gene was successfully performed on 231 evaluable isolates, however, only 9 (3.9%) showed nonsynonymous gene variants in the propeller domain of pfk13 with low, moderate, or high confidence. The Thr474Ala variant was the most prevalent, seen in six individuals, and Cys580Tyr was identified at low confidence in a single isolate of an asymptomatic individual from South Nias. There was no association of these genotypes with reduced parasite clearance, as no evidence of slow clearance by quantitative PCR (qPCR) during the first 72h following treatment with ACT.93 In 2018, a study assessing the presence of P. falciparum ACT-related resistance gene PfRBP9 showed a low frequency of the resistant allele in West Lampung.107 In 2019, pfk13 mutation was found in 15.4% of P. falciparum-infected samples in Lampung. Sequencing results identified polymorphism in position G453W, V454C, and E455K of pfk13. Mutations at codons G453W, V454C, and E455K are consistent with a recent study that found mutations at codons 440 and above in the propeller domain of the K13 gene,56 are associated with an increase in parasite clearance half-life.108 In North Sumatra, mutations in the pfk13 was identified in 4.3% of the isolates, while the rest still harbored the wild-type K13.93
In Southeast Asia, lower ART sensitivity has been linked to the pfk13 propeller domain polymorphisms. It has been independently developed in Cambodia and Myanmar and linked to P. falciparum resistance to ACT therapy.56,109,110 The prevalence of K13 mutations was extraordinarily high in western Cambodia and it increased from 40% in 2007 to 84% in 2013.109 But in the South Pacific and Southeast Asia, no incidences of P. vivax resistance to ACT have yet been documented. Despite this, numerous studies have searched for polymorphisms in the pvk12 (P. vivax ortholog of pfk13) that might result in ART resistance.111
Conclusion
For decades, reports have documented Indonesia’s history of antimalarial resistance, mainly for P. vivax and P. falciparum, starting from resistance to chloroquine in the early 1974 to sulfadoxine-pyrimethamine in 1979, which rapidly spread to many areas in Indonesia. The only antimalarial drug currently available that is still effective against both Plasmodium species is ACT, which poses a major challenge to antimalarial drug development. In endemic regions like Indonesia, some techniques have been well developed and recommended by WHO for evaluating the effectiveness of antimalarial drugs clinically and keeping track of drug resistance. Identification of molecular markers is essential to understanding the mechanism of resistance to antimalarial drug, which assists clinicians to recommend the most suitable treatment and successfully combating the Plasmodium infections. The malaria elimination strategy in Indonesia, in general, has been stated in the Indonesian Ministry of Health on April 28, 2009, which issued a Decree of the Minister of Health No. 293/MENKES/SK/IV/2009 concerning the malaria elimination program in Indonesia in 2030. This decision is supported by the Ministry of Home Affairs through Letter No. 443.41/465/SJ on 8 February 2010 concerning Guidelines for Implementation of the Malaria Elimination Program in Indonesia. Operationally, the “Gebrak Malaria National Forum” was formed through Minister of Health Decree No. 131/MENKES/SK/III/2012 on March 21, 2012. Also, the Decree of the Minister of Health No. HK.01.07/MENKES/556/2019 provides the most recent national recommendations for the medical treatment of malaria in Indonesia. However, it is not identified how to completely eliminate antimalarial resistance in Indonesia, therefore it is important that current efforts focus on properly eradicating Plasmodium sp. before further resistance occurs. In the future, continuous research in antimalarial drug-resistant markers and development of antimalarial drug with new targets are required to eradicate malaria thoroughly.
Ethical Considerations
This review article is deemed not to constitute research and the ethical clearance is therefore exempted.
Acknowledgments
The authors would like to thank the Faculty of Medicine Universitas Brawijaya. This project was supported by Faculty of Medicine Universitas Brawijaya [grant number 2371/UN10.F08/PN/2022].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev. 2002;15(4):564–594. doi:10.1128/CMR.15.4.564-594.2002
2. Cox FEG. History of the discovery of the malaria parasites and their vectors. Parasit Vectors. 2010;3(1):5. doi:10.1186/1756-3305-3-5
3. Ipa M Eliminasi malaria di Indonesia begitu sulit, mengapa? [Eliminating malaria in Indonesia is so difficult, why?]; 2018. Available from: https://theconversation.com/eliminasi-malaria-di-indonesia-begitu-sulit-mengapa-92754.
4. World Health Organization. Evidence-informed action to eliminate malaria in Indonesia; 2022. Available from: https://www.who.int/about/accountability/results/who-results-report-2020-mtr/country-story/2021/indonesia.
5. Elyazar IRF, Hay SI, Baird JK. Malaria distribution, prevalence, drug resistance and control in Indonesia. Adv Parasitol. 2011;74:41–175. doi:10.1016/B978-0-12-385897-9.00002-1
6. Ministry of Health Indonesia. Informasi Malaria Tahun 2022. [Malaria Information for 2022]. Available from: https://p2pm.kemkes.go.id/publikasi/infografis/informasi-malaria-tahun-2022.
7. Ebisawa I, Fukuyama T. Chloroquine resistance of Plasmodium falciparum in West Irian and East Kalimantan. Ann Trop Med Parasitol. 1975;69(3):275–282. doi:10.1080/00034983.1975.11687011
8. Tjitra E, Gunawan S, Laihad F, et al. Evaluation of antimalarial drugs in Indonesia, 1981–1995. Indones Bull Heal Res. 1997;25(1). doi:10.22435/bpk.v25i1Mar.242
9. Kagoro FM, Barnes KI, Marsh K, et al. Mapping genetic markers of artemisinin resistance in Plasmodium falciparum malaria in Asia: a systematic review and spatiotemporal analysis. Lancet Microbe. 2022;3(3):e184–e192. doi:10.1016/S2666-5247(21)00249-4
10. Wani WA, Jameel E, Baig U, Mumtazuddin S, Hun LT. Ferroquine and its derivatives: new generation of antimalarial agents. Eur J Med Chem. 2015;101:534–551. doi:10.1016/j.ejmech.2015.07.009
11. Sitohang V, Sariwati E, Fajariyani SB, et al. Malaria elimination in Indonesia: halfway there. Lancet Glob Heal. 2018;6(6):e604–e606. doi:10.1016/S2214-109X(18)30198-0
12. World Health Organization. “Zero malaria starts with me”: history of malaria elimination in Indonesia helps to shape a malaria-free future; 2020. Available from: https://www.who.int/indonesia/news/feature-stories/detail/zero-malaria-starts-with-me-history-of-malaria-elimination-in-indonesia-helps-to-shape-a-malaria-free-future.
13. World Heal Organisation. Word Malaria Report 2021. Geneva: World Health Organization; 2021.
14. Rahmasari FV, Asih PBS, Dewayanti FK, et al. Drug resistance of Plasmodium falciparum and Plasmodium vivax isolates in Indonesia. Malar J. 2022;21(1):1–32. doi:10.1186/s12936-022-04385-2
15. Reyburn H. New WHO guidelines for the treatment of malaria. BMJ. 2010;340(7765):161–162. doi:10.1136/BMJ.C2637
16. Kumar S, Bhardwaj TR, Prasad DN, Singh RK. Drug targets for resistant malaria: historic to future perspectives. Biomed Pharmacother. 2018;104:8–27. doi:10.1016/J.BIOPHA.2018.05.009
17. Shibeshi MA, Kifle ZD, Atnafie SA. Antimalarial Drug resistance and novel targets for antimalarial drug discovery. Infect Drug Resist. 2020;13:4047. doi:10.2147/IDR.S279433
18. Haldar K, Bhattacharjee S, Safeukui I. Drug resistance in Plasmodium. Nat Rev Microbiol. 2018;16(3):156. doi:10.1038/NRMICRO.2017.161
19. Sugiarto SR, Baird JK, Singh B, Elyazar I, Davis TME. The history and current epidemiology of malaria in Kalimantan, Indonesia. Malar J. 2022;21(1):327. doi:10.1186/s12936-022-04366-5
20. Cowell AN, Winzeler EA. The genomic architecture of antimalarial drug resistance. Brief Funct Genomics. 2019;18(5):314. doi:10.1093/BFGP/ELZ008
21. Wicht KJ, Mok S, Fidock DA. Molecular mechanisms of drug resistance in plasmodium falciparum malaria. Annu Rev Microbiol. 2020;74:431. doi:10.1146/ANNUREV-MICRO-020518-115546
22. Martin RE, Shafik SH, Richards SN. Mechanisms of resistance to the partner drugs of artemisinin in the malaria parasite. Curr Opin Pharmacol. 2018;42:71–80. doi:10.1016/j.coph.2018.07.010
23. Aliyu AW, Mustaffa KMF, Yew LC, Hou LJ. Chloroquine bioconjugates and hybrid compounds: past and recent developments in combatting chloroquine resistant malaria. Trop J Pharm Res. 2021;20(12):2663–2674. doi:10.4314/tjpr.v20i12.29
24. Cheema HS, Singh MP. Drug resistance in plasmodium, future malaria management strategies and importance of medicinal plants. J Ayurvedic Herb Med. 2022;8(2):107–112. doi:10.31254/jahm.2022.8209
25. Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185(3):380–388. doi:10.1086/338566
26. Moss S, Mańko E, Krishna S, Campino S, Clark TG, Last A. How has mass drug administration with dihydroartemisinin-piperaquine impacted molecular markers of drug resistance? A systematic review. Malar J. 2022;21(1):1–23. doi:10.1186/s12936-022-04181-y
27. Asih PBS, Rozi IE, Dewayanti FK, et al. Efficacy and safety of dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in Papua and Sumatra, Indonesia. Malar J. 2022;21(1):95. doi:10.1186/s12936-022-04101-0
28. Popovici J, Tebben K, Witkowski B, Serre D. Primaquine for Plasmodium vivax radical cure: what we do not know and why it matters. Int J Parasitol Drugs Drug Resist. 2021;15:36–42. doi:10.1016/j.ijpddr.2020.12.004
29. Camarda G, Jirawatcharadech P, Priestley RS, et al. Antimalarial activity of primaquine operates via a two-step biochemical relay. Nat Commun. 2019;10(1):3226. doi:10.1038/s41467-019-11239-0
30. Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 Alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther. 2017;102(4):688–700. doi:10.1002/cpt.690
31. Bennett JW, Pybus BS, Yadava A, et al. Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N Engl J Med. 2013;369(14):1381–1382. doi:10.1056/NEJMc1301936
32. Wang J, Zhang C-J, Chia WN, et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun. 2015;6:10111. doi:10.1038/ncomms10111
33. Ismail HM, Barton V, Phanchana M, et al. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc Natl Acad Sci U S A. 2016;113(8):2080–2085. doi:10.1073/pnas.1600459113
34. Lacy MD, Maguire JD, Barcus MJ, et al. Atovaquone/proguanil therapy for plasmodium falciparum and plasmodium vivax malaria in Indonesians who lack clinical immunity. Clin Infect Dis. 2002;35(9):e92–e95. doi:10.1086/343750
35. Ippolito MM, Moser KA, Kabuya J-B-B, Cunningham C, Juliano JJ. Antimalarial drug resistance and implications for the WHO global technical strategy. Curr Epidemiol Reports. 2021;8(2):46–62. doi:10.1007/s40471-021-00266-5
36. Bazie VB, Ouattara AK, Sagna T, et al. Resistance of plasmodium falciparum to sulfadoxine-pyrimethamine (dhfr and dhps) and artemisinin and its derivatives (K13): a major challenge for malaria elimination in West Africa. J Biosci Med. 2020;8(2):82–95. doi:10.4236/jbm.2020.82007
37. Heinberg A, Kirkman L. The molecular basis of antifolate resistance in Plasmodium falciparum: looking beyond point mutations. Ann N Y Acad Sci. 2015;1342(1):10–18. doi:10.1111/nyas.12662
38. Blasco B, Leroy D, Fidock DA. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med. 2017;23(8):917. doi:10.1038/NM.4381
39. Sanchez CP, Liu CH, Mayer S, et al. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in plasmodium falciparum. PLOS Genet. 2014;10(5):e1004382. doi:10.1371/JOURNAL.PGEN.1004382
40. Ferdig MT, Cooper RA, Mu J, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52(4):985–997. doi:10.1111/J.1365-2958.2004.04035.X
41. Cooper RA, Lane KD, Deng B, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol. 2007;63(1):270–282. doi:10.1111/J.1365-2958.2006.05511.X
42. Sidhu ABS, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194(4):528. doi:10.1086/507115
43. Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A. 1994;91(3):1143. doi:10.1073/PNAS.91.3.1143
44. Rosenthal PJ. Azithromycin for Malaria? Am J Trop Med Hyg. 2016;95(1):2. doi:10.4269/AJTMH.16-0332
45. Lell B, Kremsner PG. Clindamycin as an antimalarial drug: review of clinical trials. Antimicrob Agents Chemother. 2002;46(8):2315. doi:10.1128/AAC.46.8.2315-2320.2002
46. Gaillard T, Madamet M, Pradines B. Tetracyclines in malaria. Malar J. 2015;14(1):445. doi:10.1186/S12936-015-0980-0
47. Newton PN, Chaulet JF, Brockman A, et al. Pharmacokinetics of oral doxycycline during combination treatment of severe falciparum malaria. Antimicrob Agents Chemother. 2005;49(4):1622–1625. doi:10.1128/AAC.49.4.1622-1625.2005
48. Sheridan CM, Garcia VE, Ahyong V, DeRisi JL. The Plasmodium falciparum cytoplasmic translation apparatus: a promising therapeutic target not yet exploited by clinically approved anti-malarials. Malar J. 2018;17(1):1–13. doi:10.1186/S12936-018-2616-7
49. Dahl EL, Rosenthal PJ. multiple antibiotics exert delayed effects against the plasmodium falciparum apicoplast. Antimicrob Agents Chemother. 2007;51(10):3485. doi:10.1128/AAC.00527-07
50. Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24(6):279–284. doi:10.1016/j.pt.2008.03.007
51. Dharia NV, Plouffe D, Bopp SER, et al. Genome scanning of Amazonian Plasmodium falciparum shows subtelomeric instability and clindamycin-resistant parasites. Genome Res. 2010;20(11):1534. doi:10.1101/GR.105163.110
52. Eckstein-Ludwig U, Webb RJ, Van Goethem IDA, et al. Artemisinins target the SERCA of Plasmodium falciparum. Nat. 2003;424(6951):957–961. doi:10.1038/nature01813
53. Tilley L, Straimer J, Gnädig NF, Ralph SA, Fidock DA. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 2016;32(9):682. doi:10.1016/J.PT.2016.05.010
54. Auparakkitanon S, Wilairat P, Wilairat P. Will the in situ activator(s) of artemisinin please stand up? Mol Biochem Parasitol. 2022;248:2021–2023. doi:10.1016/j.molbiopara.2022.111461
55. Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–55. doi:10.1038/NATURE12876
56. Ashley EA, Dhorda M, Fairhurst RM, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371(5):411–423. doi:10.1056/NEJMOA1314981
57. Straimer J, Gnädig NF, Witkowski B, et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347(6220):428. doi:10.1126/SCIENCE.1260867
58. Dogovski C, Xie SC, Burgio G, et al. Targeting the cell stress response of plasmodium falciparum to overcome artemisinin resistance. PLOS Biol. 2015;13(4):e1002132. doi:10.1371/JOURNAL.PBIO.1002132
59. Mbengue A, Bhattacharjee S, Pandharkar T, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683. doi:10.1038/NATURE14412
60. Behrens HM, Schmidt S, Peigney D, May J, Maïga-Ascofaré O, Spielmann T. Moderate ART resistance mutations with low fitness cost in malaria parasites from Africa. bioRxiv. 2022;2022:5.
61. Manirakiza G, Kassaza K, Taremwa IM, Bazira J, Byarugaba F. Molecular identification and anti-malarial drug resistance profile of Plasmodium falciparum from patients attending Kisoro Hospital, southwestern Uganda. Malar J. 2022;21(1):1–10. doi:10.1186/s12936-021-04023-3
62. Nsanzabana C, Djalle D, Guérin PJ, Ménard D, González IJ. Tools for surveillance of anti-malarial drug resistance: an assessment of the current landscape. Malar J. 2018;17(1):1–16. doi:10.1186/s12936-018-2185-9
63. Slater L, Betson M, Ashraf S, Sargison N, Chaudhry U. Current methods for the detection of antimalarial drug resistance in Plasmodium parasites infecting humans. Acta Trop. 2021;216:105828. doi:10.1016/j.actatropica.2021.105828
64. World Heal Organisation. Methods for Surveillance of Antimalarial Drug Efficacy. World Heal Organisation; 2009:90.
65. World Heal Organisation. Strategy to Respond to Antimalarial Drug Resistance in Africa; 2022.
66. Bloland PB. WHO/CDS/CSR/DRS/2001.4 drug resistance in malaria drug resistance in malaria; 2001. Available from: http://www.who.int/emc.
67. Lê HG, Naw H, Kang JM, et al. Molecular profiles of multiple antimalarial drug resistance markers in plasmodium falciparum and plasmodium vivax in the Mandalay region, Myanmar. Microorganisms. 2022;10:10. doi:10.3390/microorganisms10102021
68. Shaukat A, Ali Q, Connelley T, et al. Selective sweep and phylogenetic models for the emergence and spread of pyrimethamine resistance mutations in Plasmodium vivax. Infect Genet Evol. 2019;68:221–230. doi:10.1016/j.meegid.2018.12.032
69. Haanshuus CG, Mørch K, Blomberg B, et al. Assessment of malaria real-time PCR methods and application with focus on lowlevel parasitaemia. PLoS One. 2019;14(7):1–15. doi:10.1371/journal.pone.0218982
70. Rumans LW, Dennis DT, Atmosoedjono S. Fansidar resistant falciparum malaria in Indonesia. Lancet. 1979;2(8142):580–581. doi:10.1016/S0140-6736(79)91633-7
71. Ratcliff A, Siswantoro H, Kenangalem E, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and P. vivax to chloroquine and sulfadoxine-pyrimethamine in southern Papua, Indonesia. Trans R Soc Trop Med Hyg. 2007;101(4):351–359. doi:10.1016/J.TRSTMH.2006.06.008
72. Asih PBS, Syafruddin D, Leake J, et al. Phenotyping clinical resistance to chloroquine in Plasmodium vivax in northeastern Papua, Indonesia. Int J Parasitol Drugs Drug Resist. 2011;1(1):28–32. doi:10.1016/j.ijpddr.2011.08.001
73. Fitriah F, Sulistyawati S, Riyanto S, et al. Polymorphism of Plasmodium Falciparum Dihydrofolate Reductase and Dihydropteroate Synthase Genes among Pregnant Women with Falciparum Malaria in Banjar District, South Kalimantan Province, Indonesia. J Trop Life Sci. 2012;3:92–98.
74. Reteng P, Vrisca V, Sukarno I, et al. Genetic polymorphisms in Plasmodium falciparum chloroquine resistance genes, pfcrt and pfmdr1, in North Sulawesi, Indonesia. BMC Res Notes. 2017;10(1):1–8. doi:10.1186/s13104-017-2468-1
75. Packard RM. The origins of antimalarial-drug resistance. N Engl J Med. 2014;371(5):397–399. doi:10.1056/NEJMP1403340/SUPPL_FILE/NEJMP1403340_DISCLOSURES.PDF
76. Centers for Disease Control and Prevention. Malaria - Chapter 4 - CDC Yellow Book 2020: Health Information for International Travel. Oxford University Press; 2019. Available from: https://wwwnc.cdc.gov/travel/yellowbook/2020/travel-related-infectious-diseases/malaria.
77. Lederman ER, Sutanto I, Wibudi A, et al. Imported Malaria in Jakarta, Indonesia: passive Surveillance of Returned Travelers and Military Members Postdeployment. J Travel Med. 2006;13(3):153–160. doi:10.1111/j.1708-8305.2006.00034.x
78. Ebisawa I, Fukuyama T, Kawamura Y. Additional foci of chloroquine-resistant falciparum malaria in East Kalimantan and West Irian, Indonesia. Trop Geogr Med. 1976;28:349–354.
79. Verdrager J. Effect of single dose of minocycline on a chloroquine resistant falciparum infection from balikpapan, kalimantan. Bul Penelit Kesehat. 1975;3(2):63798.
80. Tjitra E, Marwoto HA, Renny M, Ompusunggu S, Tuti S. Penelitian Obat Anti Malaria. [Anti-Malaria Drug Research]. Bul Penelit Kesehat. 1991;19(4Des). doi:10.22435/BPK.V19I4
81. Pribadi W. In vitro sensitivity of Plasmodium falciparum to chloroquine and other antimalarials in East Timor and east Kalimantan, Indonesia. Southeast Asian J Trop Med Public Heal. 1992;23:143–148.
82. Baird JK, Basri H, Subianto B, et al. Treatment of chloroquine-resistant Plasmodium vivax with chloroquine and primaquine or halofantrine. J Infect Dis. 1995;171(6):1678–1682. doi:10.1093/INFDIS/171.6.1678
83. Baird JK, Wiady I, Fryauff DJ, et al. In vivo resistance to chloroquine by Plasmodium vivax and Plasmodium falciparum at Nabire, Irian Jaya, Indonesia. Am J Trop Med Hyg. 1997;56(6):627–631. doi:10.4269/AJTMH.1997.56.627
84. Sumawinata IW, Bernadeta B. Very high risk of therapeutic failure with chloroquine for uncomplicated Plasmodium falciparum and P. vivax malaria in Indonesian Papua - PubMed. Am J Trop Med Hyg. 2003;68(4):416–420.
85. Fryauff DJ, Soekartono TS. Survey of resistance in vivo to chloroquine of Plasmodium falciparum and P. vivax in North Sulawesi, Indonesia. Trans R Soc Trop Med Hyg. 1998;92(1):82–83. doi:10.1016/s0035-9203(98)90966-x
86. Sutanto I, Suprijanto S, Nurhayati MP, Baird JK. Resistance to chloroquine by Plasmodium vivax at Alor in the Lesser Sundas Archipelago in eastern Indonesia. Am J Trop Med Hyg. 2009;81(2):338–342.
87. Sutanto I, Endawati D, Ling LH, Laihad F, Setiabudy R, Baird JK. Evaluation of chloroquine therapy for vivax and falciparum malaria in southern Sumatra, western Indonesia. Malar J. 2010;9(1):11–15. doi:10.1186/1475-2875-9-52
88. Suwanarusk R, Russell B, Chavchich M, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One. 2007;2(10):e1089. doi:10.1371/journal.pone.0001089
89. Siswantoro H, Ratcliff A, Kenangalem E, et al. Efficacy of existing antimalarial drugs for uncomplicated malaria in Timika, Papua, Indonesia. Med J Indones. 2006;15(4):251–258. doi:10.13181/mji.v15i4.243
90. Asih PB, Rogers WO, Susanti AI, et al. Seasonal distribution of anti-malarial drug resistance alleles on the island of Sumba, Indonesia. Malar J. 2009;8(1):1–7. doi:10.1186/1475-2875-8-222
91. Salwati E, Herman R, Handayani S, Tjitra E, Deteksi P. Vivax Single Nucleotide Polymorphism (SNP) Y976F dari Sampel Monitoring Pengobatan Dihidroartemisinin-Piperakuin di Kalimantan dan Sulawesi. [Detection of P.vivax Single Nucleotide Polymorphism (SNP) Y976F from Monitoring Samples of Dihydroartemi. Media Penelit Dan Pengemb Kesehat. 2012;22. doi:10.22435/MPK.V22I3
92. Saleh I, Handayani D, Anwar C. Polymorphisms in the pfcrt and pfmdr1 genes in Plasmodium falciparum isolates from South Sumatera, Indonesia. Med J Indones. 2014;23(1):3–8. doi:10.13181/MJI.V23I1.679
93. Lubis IND, Wijaya H, Lubis M, Lubi CP, Beshi KB, Sutherlan CJ. Plasmodium falciparum Isolates Carrying pfk13 Polymorphisms Harbor the SVMNT Allele of pfcrt in Northwestern Indonesia. Antimicrob Agents Chemother. 2020;64:8. doi:10.1128/AAC.02539-19
94. Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47(8):2418–2423. doi:10.1128/AAC.47.8.2418-2423.2003
95. Ross LS, Dhingra SK, Mok S, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun. 2018;9(1):25–28. doi:10.1038/s41467-018-05652-0
96. Calçada C, Silva M, Baptista V, et al. Expansion of a Specific Plasmodium falciparum PfMDR1 Haplotype in Southeast Asia with Increased Substrate Transport. MBio. 2020;11:6. doi:10.1128/mBio.02093-20
97. Nagesha HS. Mutations in the pfmdr1, dhfr and dhps genes of Plasmodium falciparum are associated with in-vivo drug resistance in West Papua, Indonesia. Trans R Soc Trop Med Hyg. 2001;95(1):43–49. doi:10.1016/s0035-9203(01)90329-3
98. Hastings MD, Porter KM, Maguire JD, et al. Dihydrofolate reductase mutations in plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J Infect Dis. 2004;189(4):744–750. doi:10.1086/381397
99. Syafruddin D, Asih PBS, Casey GJ, et al. Molecular epidemiology of Plasmodium falciparum resistance to antimalarial drugs in Indonesia. Am J Trop Med Hyg. 2005;72(2):174–181. doi:10.4269/ajtmh.2005.72.174
100. Maguire JD, Lacy MD, Sururi H, et al. Chloroquine or sulfadoxine-pyrimethamine for the treatment of uncomplicated, Plasmodium falciparum malaria during an epidemic in Central Java, Indonesia. Ann Trop Med Parasitol. 2002;96(7):655–668. doi:10.1179/000349802125002310
101. Fryauff DJ, Leksana B, Masbar S, et al. The drug sensitivity and transmission dynamics of human malaria on Nias Island, North Sumatra, Indonesia. Ann Trop Med Parasitol. 2002;96(5):447–462. doi:10.1179/000349802125001249
102. Basuki S, Fitriah RS, Budiono DYP, Uemura H. Two novel mutations of pfdhps K540T and I588F, affecting sulphadoxine-pyrimethamine-resistant response in uncomplicated falciparum malaria at Banjar district, South Kalimantan Province, Indonesia. Malar J. 2014;13(1):135. doi:10.1186/1475-2875-13-135
103. Asih PBS, Marantina SS, Nababan R, et al. Distribution of Plasmodium vivax pvdhfr and pvdhps alleles and their association with sulfadoxine–pyrimethamine treatment outcomes in Indonesia. Malar J. 2015;14(1):365. doi:10.1186/s12936-015-0903-0
104. Jiang T, Cheng W, Yao Y, Tan H, Wu K, Li J. Molecular surveillance of anti-malarial resistance Pfdhfr and Pfdhps polymorphisms in African and Southeast Asia Plasmodium falciparum imported parasites to Wuhan, China. Malar J. 2020;19(1):434. doi:10.1186/s12936-020-03509-w
105. Kaingona-Daniel EPS, Gomes LR, Gama BE, et al. Low-grade sulfadoxine-pyrimethamine resistance in Plasmodium falciparum parasites from Lubango, Angola. Malar J. 2016;15:309. doi:10.1186/s12936-016-1358-7
106. Asih PBS, Dewi RM, Tuti S, et al. Efficacy of Artemisinin-Based Combination Therapy for Treatment of Persons with Uncomplicated Plasmodium falciparum Malaria in West Sumba District, East Nusa Tenggara Province, Indonesia, and Genotypic Profiles of the Parasite. Am J Trop Med Hyg. 2009;80(6):914–918. doi:10.4269/AJTMH.2009.80.914
107. Ghiffari A, Rahman R, Anwar C. No antimalarial resistance of Plasmodium falciparum varian West Indonesia using molecular markers screening. J Phys Conf Ser. 2019;1282:1. doi:10.1088/1742-6596/1282/1/012077
108. Rachmad B. Isolasi dan Identifikasi Mutasi Gen PfK13 (PF3D7_1343700) sebagai penanda resistensi artemisinin pada isolat plasmodium falciparum Asal lampung. [Isolation and Identification of PfK13 gene mutation (PF3D7_1343700) as a marker of artemisinin resistance in P. Pros dalam rangka Rakernas. 2019;2019:1.
109. Amato R, Pearson RD, Almagro-Garcia J, et al. Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: a retrospective genetic study. Lancet Infect Dis. 2018;18(3):337–345. doi:10.1016/S1473-3099(18)30068-9
110. Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364(9432):438–447. doi:10.1016/S0140-6736(04)16767-6
111. Sidhu AB, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57(4):913–26. doi:10.1111/j.1365-2958.2005.04729.x
112. Sisowath C, Strömberg J, Mårtensson A, et al. In Vivo Selection of Plasmodium falciparum pfmdr1 86N Coding Alleles by Artemether‐Lumefantrine (Coartem). J Infect Dis. 2005;191(6):1014–1017. doi:10.1086/427997
113. Sisowath C, Ferreira PE, Bustamante LY, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Tropical Medicine & International Health. 2007;12(6):736–742. doi:10.1111/j.1365-3156.2007.01843.x
114. Lu K, Derbyshire ER. Tafenoquine: A Step toward Malaria Elimination. Biochemistry. 2020;59(8):911–920. doi:10.1021/acs.biochem.9b01105
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.