Back to Journals » Veterinary Medicine: Research and Reports » Volume 12
Antibiotic Resistance Development in Animal Production: A Cross-Sectional Study
Authors Gebeyehu DT
Received 9 March 2021
Accepted for publication 28 April 2021
Published 18 May 2021 Volume 2021:12 Pages 101—108
DOI https://doi.org/10.2147/VMRR.S310169
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Young Lyoo
Daniel Teshome Gebeyehu
Department of Veterinary Medicine, School of Veterinary Medicine, Wollo University, Dessie, Ethiopia
Correspondence: Daniel Teshome Gebeyehu Email [email protected]
Background: In recent years, an increase in the development of antimicrobial-resistant pathogens especially foodborne zoonotic bacteria has been observed. As a result, crude mortality rates are increasing due to those resistant bacteria in both human and animal populations, particularly in developing countries like Tanzania where the risk of infection is high due to poor biosecurity measures, close animal–human interactions, and extensive use of antimicrobials for animal productions. One of those zoonotic bacterial pathogens, which commonly contaminates food, is Salmonella.
Methodology: A cross-sectional study was carried out on samples collected from diarrheic sheep, to assess the level of antibiotic resistance of Salmonella. From 165 fecal samples, 80 of which were tested positive for Salmonella. The antibiotic resistance level of Salmonella isolates was conducted by the Kirby-Bauer disc diffusion method using ten commonly used antibiotics in the study area.
Results: Out of 80 Salmonella positive samples, all (100%) of them were resistant to amoxicillin and ampicillin while sixty-eight (85%), 68 (85%), and 60 (75%) isolates were susceptible to gentamicin, ciprofloxacin, and kanamycin, respectively. Thirty (37.5%) Salmonella isolates were resistant to both trimethoprim and tetracycline and 25% of the isolates were resistant to both doxycycline and chloramphenicol while 12.5% of the isolates were resistant to nalidixic acid.
Conclusion: This study revealed that the Salmonella isolates of diarrheic sheep developed a wide range of resistance to different antibiotics. Further studies and integrative approaches in a one health framework among animal–human and environmental health professionals are recommended for the mitigation of health risks arising from antibiotic-resistant zoonotic pathogens like Salmonella.
Keywords: food animals, isolation, Salmonella, antibiotics, antibiotic resistance, sheep
Introduction
Although antimicrobials contribute a vital role in the treatment and prophylactic activities, they are losing their effectiveness due to the dramatic increase of antimicrobial resistance (AMR). Even though different factors aggravate AMR, overuse, and misuse of antimicrobials in other sectors play a significant role. Among the activities that increase AMR: using antibiotics in animal production, using drugs without professional oversight, and poor diagnostic techniques.1,2 Since it affects both the animal and human health sectors, the risk of AMR in zoonotic diseases is severe than the risk of AMR in non-zoonotic diseases. People in developing countries, living in poor hygienic conditions and consuming raw animal origin food are at a high risk of affecting by antimicrobial-resistant zoonotic diseases.1
Like other bacteria, Salmonella is developing resistance against antibiotics based on either of the following resistance mechanisms: production of enzymes that inactivate antimicrobial agents through degradation or structural modification, reduction of bacterial cell permeability to antibiotics, activation of antimicrobial efflux pumps, and modification of the cellular target for the drug.3,4
The resistance of Salmonella against commonly used and previously curative antibiotics is dramatically increasing with broad geographic coverage.5–9 The mortalities of patients, who were contracted by antibiotic-resistant Salmonella are high and the economic loss due to morbidity is increasing as well.10 The Zoonotic nature of Salmonella makes the AMR against it worst, in which it can be transmitted easily from animal to human.11–13 The exposure of Salmonella to different antibiotics, while it is in the environment or inside animals, enables it to develop resistance. The resistant Salmonella then quickly pass to human through the food chain and cause huge health crises.14
Even if the international tripartite organizations (WHO, FAO, and OIE) and Codex Alimentarius are working against antibiotic resistance and set action plans to minimize antibiotic use along the food chain, the use of antibiotics is increasing at an alarming rate.15,16 Before this research, many other types of research were conducted to assess the resistance pattern of Salmonella both in human and animals, but due to the “evolutionary nature of the bacteria and the dependency of patients to antibiotics”, the resistance level of Salmonella to antibiotics has been increasing.6–9,17–20 Antibiotic resistance surveillance of foodborne pathogens in food animals like sheep is strongly recommended to mitigate the global crises due to AMR and there was no surveillance had been done yet in the study area.
Isolation of the zoonotic disease-causing agent (like Salmonella) and conducting antibiotic sensitivity testing are the recommended activities to see the antibiotic resistance phenomenon of zoonotic pathogens in animals.21 Such surveillance types are the foundation for conducting further studies, formulating antimicrobial use policies, and creating public awareness.
Therefore, the objective of this study was to assess the antibiotic resistance level of Salmonella isolated from diarrheic sheep.
Materials and Methods
Study Area
The study was conducted in and around Morogoro city, Tanzania. The climate condition of Morogoro is hot with an average temperature of 28 °C and an annual rainfall of 889mm. It elevates 509m above sea level with a latitude and longitude of 6.8278° S and 37.6591° E respectively. Morogoro city is located 263km away from Dodoma (the Capital city of Tanzania). In the area, there is a large number of livestock populations reared in the different management systems, and antibiotics were massively used for different purposes (prophylaxis, metaphylaxis, disease treatment, and growth promotion) in animal production.
Study Population
The antibiotic resistance test was performed on Salmonella isolates, collected from sheep in veterinary clinics of the study area, during the outbreak of diarrheal disease. All diarrheic sheep were sampled and Salmonella was isolated from the feces of those sheep. Since the owners were not agreed, some sheep were excluded from sampling.
Study Design
A cross-sectional study was conducted from September 2019 to March 2020. There is no scientific reason to select this study time, but the diarrheal outbreak occurred at that time and the researcher was motivated to conduct the study by his own initiative. The result of antibiotic resistance tests was used to understand the progress of antibiotic resistance in zoonotic disease-causing Salmonella in sheep.
Sample Collection Techniques and Antibiotic Resistance Testing Procedure
Sample Collection Techniques
Diarrheic fecal samples were collected from 165 sheep from five veterinary clinics in and around Morogoro city. The swab samples were placed in test tubes filled with peptone water and leveled with special IDs for each sample. The test tubes that contain samples were placed an icebox until it reaches the Sokoine University of Agriculture microbiology laboratory. After the samples arrive in this laboratory it was stored in 4°C refrigerator for a night and the culturing and isolation procedures were done in the next day. After consecutive culture and biochemical characterization processes, 80 samples were positive for Salmonella.
Isolation and Identification Process of Salmonella
Isolation of Salmonella was performed as recommended by the American food and drug administration.22,23 In brief, samples were pre-enriched into 9 mL of buffered peptone water and incubated at 37°C for 18 hours. Aseptically 100µL of the pre-enriched inoculums were transferred to 10 mL of Rappaport-Vassiliadis broth and incubated at 42°C for 24 hours. Positive enrichment Rappaport Vassiliadis tubes were inoculated onto MacConkey Agar. MacConkey Agar (MAC) is a selective and differential medium designed to isolate and differentiate enteric bacteria based on their ability to ferment lactose and the inoculated plates were incubated at 37°C for 24 hours.
Gram’s staining was performed on a pure colony24 to determine the shape and arrangement of bacteria. The non-lactose fermenting colorless colonies from the MacConkey Agar (Salmonella) were subcultured onto xylose lysine desoxycholate (XLD) agar and incubated at 37°C for 24 hours for the appearance of the characteristic colorless colony with a black center. The presumptive Salmonella colony appearing slightly transparent red halo with a black center surrounded by a pink-red zone on XLD agar were screened, further for its biochemical characterization. Standard biochemical tests, Catalase test, Indole, Methyl red, Voges-Proskauer test, Citrate utilization, urease test, Triple sugar Iron test, and Carbohydrate fermentation test were used as confirmation of identification.25,26
Preparation of 0.5 McFarland Standard
According to,19 0.5mL of BaCl2 (1.175% BaCl2.2H2O) was added to 99.5 mL of H2SO4 (1%), and the standard solution was poured into plastic capped tubes of the same size and volume as those used in the bacterial suspension. The tubes were tightly sealed to prevent loss by evaporation and stored protected from light at room temperature (20 °C) and the standard was vigorously agitated in a vortex mixer before use.
Antimicrobial Resistance Testing of Salmonella
The isolated Salmonella was screened for an in-vitro antimicrobial resistance test using the agar disk diffusion method.27 Antibiotic resistance tests of Salmonella isolates were done on the commonly used antibiotics in the study area for the purpose of animal production. Ten different antibiotics (OXOID, England) discs with their concentrations given in parentheses were used in the antibiograms are kanamycin (K, 30µg), gentamicin (CN, 10µg), amoxicillin (AMX, 10µg), doxycycline (DO, 30µg), chloramphenicol (C, 30µg), tetracycline (TE, 30µg), ciprofloxacin (CIP, 5µg), trimethoprim (TMP, 5µg), nalidixic acid (NAL, 30µg) and ampicillin (AMP, 10µg). Since the aim of this study was to test the curative efficiency of commonly used antibiotics in the study area, the dose of the antibiotic disc which has a similar antibiotic dose with the locally used antibiotics was chosen to test the antibiotic resistance level of Salmonella isolates.
An isolated colony from nutrient agar plates was transferred by sterilized inoculating loop into tubes containing 5 mL of the normal saline solution until it achieved 0.5 McFarland turbidity standards and then a sterile cotton swab was dipped into the adjusted suspension and pressed against the inside of the tube to remove excess water. The swab was then spread evenly over the entire surface of the plate of Mueller-Hinton agar (OXOID) to obtain uniform inoculums. The plates were allowed to dry for 5 minutes. Antibiotics impregnated discs were then applied to the surface of the inoculated plates with a disc dispenser. Each disc was gently pressed down onto the agar by sterile forceps to ensure complete contact with the agar surface.
Even distribution of discs and minimum distance of 24 mm from the center to center and from the edge of the plate was ensured to prevent overlapping of the inhibition zones. Four antibiotic discs were placed in each Petri dish. Within 15 minutes of the application of the discs, the plates were incubated at 37°C. After 24 hours of incubation, the plates were examined, and the diameter of the zones of complete inhibition to the nearest whole millimeter was measured by a digital caliper. Based on the Clinical and Laboratory Standard Institute,28 the clear zones (inhibition zones) around the antibiotic discs (including the disc) were expressed as Susceptible (S), Intermediate (I), and Resistant (R) (Table 1).
 |
Table 1 Comparison of the Inhibition Zone of the Standard with the Average Finding of the Present Study and Standard Deviations of the Inhibition Zones |
Ethics Statement
The fecal samples were collected from sheep that came to the veterinary clinics for the treatment of diarrheal cases in the study area. The researchers asked the agreement of the sheep owners to take fecal samples from their sheep for research purposes. The agreement was verbal agreement. For the owners who was not volunteer, samples were not taken from their animals. Confidentiality of the sample and all other sources were maintained by the research ethics guidelines of the university. So, in this study animals, humans, or tissue from animal or human was not involved directly, but only feces.
Data Analysis
The dataset for analysis was prepared using Microsoft excel 2013 and descriptive statistics (graphs, numbers, tables, and percentages) were used to describe findings. Each antibiotic was tested for all 80 isolated Salmonella. The average inhibition zones of all 80 tests of each antibiotic were calculated and one single inhibition zone for each antibiotic was produced, to ease analysis. One sample can be resistant to more than one antibiotic. The nature of resistance (multiple or single) and the number of resistant samples for each antibiotic are presented in tables.
Result
During the study, 165 total samples were examined. After consecutive culture and biochemical test activities, 80 of them were tested positive for Salmonella. The resistance level of selected antibiotics was tested on each isolate. As the antibiotic sensitivity test result revealed, Salmonella isolates were completely (100%) resistant to ampicillin and amoxicillin, whereas 70 (87.5%) isolates were susceptible to gentamicin and ciprofloxacin. Also, 60 (75%) of Salmonella isolates were susceptible to kanamycin and chloramphenicol while 40 (50%), 30 (37.5%), 30 (37.5%), and 20 (25%) of the isolates were resistant to nalidixic acid, trimethoprim, tetracycline, and doxycycline respectively (Figure 1).
 |
Figure 1 Overall antibiotic resistance level of Salmonella isolates against selected antibiotics in percent (%). |
According to the clinical and laboratory standard institute (CLSI),28 antibiotic sensitivities were expressed in three modalities (sensitive, intermediate, and resistance). To categorize the sensitivity of Salmonella into these three modalities, CLSI uses the measurement results of the inhibition zone in millimeters. The average inhibition zone of all 80 Salmonella positive samples by comparing them with the standard and the standard deviation of the inhibition zones are indicated in Table 1.
All (80) Salmonella isolates were developed resistance to more than one antibiotic (multidrug-resistant). All (100%) of the isolates were resistant to both ampicillin and amoxicillin, whereas all isolates were fully or intermediately susceptible to 3 antibiotics (gentamicin, kanamycin, and ciprofloxacin). The details of multidrug resistance are shown in Table 2.
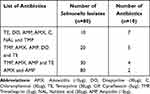 |
Table 2 The Number of Salmonella Isolates That Was Developed Multi-Drug Resistance |
Discussion
All (80) Salmonella isolates found multidrug-resistant. In agreement with this study, the zoonotic Salmonella isolates were developed multidrug resistance in Modjo,20 Gondar,7 Jimma,9 and China,8 Morocco.29 This multidrug resistance might be attributed to the irrational use of antibiotics practiced in the area30,31 and/or the evolutionary nature of Salmonella species.18 The multidrug resistance bacteria causes a dangerous health crisis than resistance against single antibiotics, especially the crisis is very bad when it is on zoonotic diseases. Since it easily circulates between animals and humans, it is difficult to eradicate zoonotic resistance bacteria.5 As a result, one health approach is highly necessitating for proper mitigation of these diseases.
Salmonella isolates of the current study were sensitive to gentamicin, kanamycin, and ciprofloxacin. Likewise, zoonotic Salmonella isolated from diarrheic sheep in San Angelo were 100% sensitive to ciprofloxacin and gentamicin.32 As reported by the same study, 99.37% of Salmonella isolates were sensitive to kanamycin.32 Specifically, the sensitivity of zoonotic Salmonella to gentamicin agrees with the study conducted in Harar33 with a 92.8% sensitivity level. In addition to this, the study done by Raseta et al34 in Indonesia revealed that 92% of zoonotic Salmonella isolates were sensitive to gentamicin. The present finding also slightly agrees with the study done in Addis Ababa by Mamuye et al35 with the samples collected from diarrheic children, which was 50% sensitive, 50% intermediate, and 0% resistant to both gentamicin and kanamycin. The rationale of comparing the prevalence of superbug zoonotic Salmonella in diarrheic sheep and diarrheic children is that the high prevalence of this antibiotic-resistant zoonotic pathogen is the problem of both human and animal health sectors regardless of where the resistance is developed. This slight variation of antibiotic sensitivity might be attributed to the difference in the level of antimicrobial use in human and animal health sectors. Standing from the result of the present study, prudent use of these effective antibiotics is recommended to preserve them as a choice of treatment for the future.
In contrast to the current finding, a high level of resistance to gentamicin (75.6%) was reported by.35 In the same study area from samples collected from human diarrhea and a study done by Brooks et al36 resistance in Kenya with a resistance level of 13.0% to kanamycin. Resistance to gentamicin which was reported by35,37 might be due to extensive use of gentamicin in human medicine compared with veterinary medicine.38 The difference can be attributed to the differences in geographical locations, antimicrobial use policies, and study time. As described by the joint concept note of FAO-OIE-WHO,16 an integrated antimicrobial use policy is the most determinant factor for the development of antimicrobial resistance. As a result, if there is no antimicrobial use policy/regulation, there will be unlimited antimicrobial use in both animal and human health sectors, which in turn aggravates antimicrobial resistance.
Salmonella isolates were fully developed resistance to amoxicillin and ampicillin. The current results were consistent with those revealed from studies in Harer33 and West Showa,39 in which Salmonella isolates were 100% resistant for both ampicillin and amoxicillin. In the researches done in Nekemt40 and Addis Ababa,36 Salmonella showed 90 and 80% resistance respectively to amoxicillin. Comparable to the present results, Salmonella was 90% resistant to ampicillin in Taiwan.41 In addition to these reports, resistant Salmonella was most frequent to amoxicillin reported in Serbia42 and to ampicillin in Nigeria.43
Unlike the result of the current result, ampicillin, and amoxicillin were effective in Salmonella in the same study area conduct in human hospitals.36 The difference between these findings can be due to the difference in the study time, animal and human health professionals’ practices, and extensive use of antibiotics for prophylaxis, anaphylaxis, and metaphylaxis purposes in animal production.2
In the present study, the majority (75%) of the Salmonella isolates were susceptible to chloramphenicol with 12.5% intermediate and 12.5% resistance. Also, 50% of the isolates were susceptible to nalidixic acid with 12.5% resistance and 37.5% intermediate. Similarly, zoonotic Salmonella collected from the raw beef in Nigeria showed 19.1% resistance to chloramphenicol.44 Comparable to the present study, tetracycline revealed 42.41% sensitivity for Salmonella in the study conducted by Cetin et al,32 and on the opposite, Salmonella was highly resistant (96.2%) to nalidixic acid in the research done by the same researcher.32 In contrast to the present study, a high level of resistance to tetracycline (100%), ciprofloxacin (63.6%), and chloramphenicol (45%) were reported in Nigeria.44 This difference might be due to differences in antimicrobial using practice, geographic location, and antimicrobial use policy.16
Similar to the present study, Salmonella isolated from raw beef in Burkina Faso showed a high level of sensitivity pattern to ciprofloxacin and chloramphenicol,42 and high level of resistance to tetracycline (95%), and a moderate level of resistance to gentamicin (37%) and nalidixic acid (32%) was reported in Nigeria.43 This might be due to the widely used practice of these drugs in the country. On the other hand, the main difference among the outcome of the studies is since there is strain variation in resistance to a given antibiotic. For instance, Salmonella enteric serovar Paratyphi-A was susceptible to gentamicin (100%), chloramphenicol (95%), and tetracycline (92.5%). Salmonella enteric serovar Typhi was susceptible to gentamicin (97.7%), chloramphenicol (97.5%) and tetracycline (93.2%).44
Among the prospective actions that the researchers recommended are the followings. Firstly, multidisciplinary collaborations among different sectors, actors, and professionals have demanded the mitigation of antimicrobial use and resistance. Secondly, awareness creation about the severity of antimicrobial-resistant zoonotic disease risk should be communicated to different stakeholders (animal producers, policymakers, and antimicrobial use regulators). Thirdly, appropriate diagnostic equipment should be furnished for reliable and informative antimicrobial resistance surveillance activities. Lastly, further antimicrobial resistance investigations followed by new drug development and facilitating alternative antimicrobial uses, like vaccination and biosecurity measures in animal production are recommended.
Conclusion
Antibiotic resistances for Salmonella isolates were high in the study area. The high level of antibiotic resistance that was seen in this study can occur as a result of irrational antimicrobial use practices (prophylaxis, anaphylaxis, metaphylaxis, and growth promotion) in sheep production. Other zoonotic disease causative agents are expected to develop the same level of resistance as Salmonella. The majority (7 out of 10) of the antibiotics were lost their curative efficiency for Salmonella isolates in the study area. In addition to ampicillin and amoxicillin, which are totally (100%) not effective for the treatment of Salmonellosis in the study area, others are on the way of losing their curative nature forever. More devastatingly, all resistant Salmonella were developed multidrug resistance. This study showed that unless and other ways we take urgent and integrated measures in a one health framework, the whole antibiotics will be useless shortly. In this rapid resistance development rate, the currently curable food animal origin zoonotic diseases will be out of control due to a lack of effective antibiotics.
Data Sharing Statement
The data used to support the findings of this study are included in the article in the frequency table.
Acknowledgment
I acknowledge the Sokoine University Veterinary Medicine teachers and laboratory workers for supporting me during this research work. Lastly, I forward my thanks to the sheep owners for allowing me to take samples from their sheep.
Funding
Since the research is done based on personal initiative, anyone didn’t fund this research work.
Disclosure
The author declares that there is no competing interest.
References
1. World Health Organization. Implementation of the global action plan on antimicrobial resistance, WHO GAP AMR Newsletter No.32; 2017. Available from: https://www.who.int/antimicrobial-resistance/en/.
2. Morar A, Sala C, Imre K. Occurrence and antimicrobial resistance of Salmonella isolates recovered from the pig slaughter process in Romania. J Infect Dev Ctries. 2015;9(1):99–104. doi:10.3855/jidc.5236
3. Foley SL, Lynne AM. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci. 2008;86(14):173–187. doi:10.2527/jas.2007-0447
4. Ibarra JA, Steele-Mortimer O. Salmonella the ultimate insider Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579–1586. doi:10.1111/j.1462-5822.2009.01368.x
5. Humphrey TJ. Public-health aspects of Salmonella infections. In: Wray C, Wray A, editors. Salmonella in Domestic Animals. Wallingford, England: CABI Publishing; 2000.
6. Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol Rev. 2002;26:141–148. doi:10.1111/j.1574-6976.2002.tb00606.x
7. Ejo M, Garedew L, Alebachew Z, et al. Prevalence and antimicrobial resistance of Salmonella isolated from animal-origin food items in Gondar, Ethiopia. Biomed Res Int. 2016. doi:10.1155/2016/4290506.
8. Wang X, Biswas S, Paudyal N, et al. Antibiotic resistance in Salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front Microbiol. 2019;10:985. doi:10.3389/fmicb.2019.0098.
9. Taddese D, Tolosa T, Deresa B, et al. Antibiograms and risk factors of Salmonella isolates from laying hens and eggs in Jimma Town, South Western Ethiopia. BMC Res Notes. 2019;12:1–7. doi:10.1186/s13104-019-4516-5.
10. Cailhol J, Lailler R, Bouvet PL, Vieille S, Gauchard F, Sanders P. Trends in antimicrobial resistance phenotypes in non-typhoid Salmonella from human and poultry origins in France. Epidemiol Infect. 2006;134:171–178. doi:10.1017/S0950268805004863
11. Varma JK, Molbak K, Barrett TJ, Beebe JL, Jones TF, Rabatsky H. Antimicrobial-resistant non typhoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis. 2005;191:554–561. doi:10.1086/427263
12. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379(9835):2489–2499. doi:10.1016/S0140-6736(11)61752-2
13. Xu X, Chen Y, Pan H, et al. Genomic characterization of Salmonella Uzaramo for human invasive infection. Microb Genom. 2020;6. doi:10.1099/mgen.0.000401
14. Liu Y, Jiang J, Ed-Dra A, et al. Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res Int. 2021;142:110198. doi:10.1016/j.foodres.2021.110198
15. Codex Alimentarius. Code of practice to minimize and contain antimicrobial resistance, CAC/RCP. 2005:61–2005.
16. FAO-OIE-WHO Collaboration. Sharing responsibilities and coordinating global activities to address health risks at the animal-human-ecosystems interfaces, “A Tripartite Concept Note”. 2016.
17. Akinyemi KO, Phillip W, Beyer W, Bohm R. In –vitro antimicrobial resistance patterns of Salmonella enteric serovars and emergence of Salmonella phage type DT071 in a suspected community-associated outbreak in Lagos, Nigeria. J Infect Dev Ctries. 2007;1(1):48–54.
18. World Health Organization. The evolving threat of antibiotic resistance: option of action. World Health Organization; 2017:p1–p119.
19. Akond MA, Shirin M, Alam S, Hassan SM, Rahman M, Hoq M. Frequency of drug resistant Salmonella spp. isolated from poultry samples in Bangladesh. Stamford J Microbiol. 2012;2(1):2074–5346. doi:10.3329/sjm.v2i1.15207
20. Xu Y, Zhou X, Jiang Z, et al. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front Cell Infect Microbiol. 2020;10. doi:10.3389/fcimb.2020.00497
21. Asfaw Ali D, Tadesse B, Ebabu A. Prevalence and antibiotic resistance pattern of Salmonella isolated from caecal contents of exotic chicken in Debre Zeit and Modjo, Ethiopia. Biomed Res Int. 2020;2020. doi:10.1155/2020/1910630
22. Andrews WH, Jacobson A, Hammack TS. Bacteriological Analytical Manual.
23. Prescott JF. Veterinary microbiology and microbial disease. Can Vet J. 2003;44(12):986.
24. Merchant IA, Packer RA. Veterinary Bacteriology and Virology.
25. Jarvis CJ, Kellerman GE, Van Rensburg WJJ, Whitehead CJ. The Bacteriology Manual.
26. Brenner DJ, Krieg NR, Staley JT. Bergey’s Manual of Systematic Bacteriology.
27. Bauer AW, Kirby M, Sheris JD, Turch M. Antibiotic resistance testing by standard single disc method. Am J Clin Pathol. 1966;45(4):493–496. doi:10.1093/ajcp/45.4_ts.493
28. CLSI. Performance standards for antimicrobial resistance testing; Twenty-fifth informational supplement. CLSI document M100-S25. Wayne, PA; 2017.
29. Ed-Dra A, Filali FR, Khayi S, et al. Antimicrobial resistance, virulence genes, and genetic diversity of Salmonella enterica isolated from sausages. Eur J Microbiol Immunol. 2019;9(2):56–61. doi:10.1556/1886.2018.00035.
30. Gilberg K, Laouri M, Wade S, Isonaka S. Analysis of medication use patterns: apparent overuse of antibiotics and underuse of prescription drugs for asthma, depression, and CHF. J Manage Care Pharm. 2003;9(3):232–237. doi:10.18553/jmcp.2003.9.3.232
31. Kasper DL, Fauci AS, Longo DL, Braunwald E, Hauser SL, Jameson JL, Eds. Harrison’s Principles of Internal Medicine. New York: The Mc Graw-Hill companies; 2005:897–906.
32. Cetin E, Temelli S, Eyigor A. Nontyphoid Salmonella prevalence, serovar distribution and antimicrobial resistance in slaughter sheep. Food Sci Anim Resour. 2020;40(1):21–33. doi:10.5851/kosfa.2019.e75
33. Ayalu AR, Berhanu S, Jemal Y, Gizachew A, Sisay F, Jean-Michel V. Antibiotic resistance patterns of Salmonella and Shigella isolates in Harar, Eastern Ethiopia. J Infect Dis Immun. 2011;3(8):134–139.
34. Raseta M, Mrdovic B, Jankovic V, et al. Prevalence and antibiotic resistance of Salmonella spp. In meat products, meat preparations and minced meat. In:
35. Mamuye Y, Metaferia G, Birhanu A, et al. isolation and antibiotic Resistance pattern of shigella and Salmonella among under 5 children with acute diarrhea. Clin Microbiol. 2015;4:186. doi:10.4172/2327-5073.1000186
36. Brooks JT, Ochieng JB, Kumar L, et al. Surveillance for bacterial diarrhea and antimicrobial resistance in rural western Kenya, 1997–2003. Clin Infect Dis. 2006;43:393–401. doi:10.1086/505866
37. European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDPC). The European Union Summary Report on antimicrobial resistance in Antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in the European Union in 2014. EFSA J. 2016;14:4380.
38. Gunasekara T, Yonas H, Asefa B. Antibiotic susceptability pattern of Salmonella and Shigela among diarrhal patients, in Gedo Hospital, West Shoa Zone, Oromia State, Ethiopia. Am J Drug Discov Dev. 2014;4(3):194–204. doi:10.3923/ajdd.2014.194.201
39. Alemayehu T, Mulissa J. Prevalance and antibiotic resistance pattern of Salmonella and shigella species among diarrheal patients attending Nekemte Referal Hospital, Oromia, Ethiopia. Int J Microbiol. 2017. doi:10.1155/20189214689
40. Angelo KM, Reynolds J, Karp BE, et al. Antimicrobial resistance among nontyphoidal Salmonella isolated from blood in the United States, 2003–2013. J Infect Dis. 2016;214(10):1565–1570. doi:10.1093/infdis/jiw415
41. Petrovica J, Babic J, Stojanova I, Velhnera M. The change in antimicrobial resistance profile of meat chain-associated Salmonella in Serbia. Procedia Food Sci. 2015;5:231–234. doi:10.1016/j.profoo.2015.09.059
42. Bawa IH, Tchamba GB, Bagré TS, et al. Antimicrobial resistance of Salmonella enterica strains isolated from raw beef, mutton and intestines sold in Ouagadougou, Burkina Faso. J Appl Biosci. 2015;95:8966–8972. doi:10.4314/jab.v95i1.6
43. Umeh SI, Enwuru CP. Antibiotic resistance profile of Salmonella isolates from livestock. Open J Medical Microbio. 2014;4:242–248. doi:10.4236/ojmm.2014.44027
44. Bata SI, Karshima NS, Yohanna J, Dashe M, Pam VA, Ogbu KI. Isolation and antibiotic sensitivity patterns of Salmonella species from raw beef and quail eggs from farms and retail outlets in Jos, Plateau State, Nigeria. J Vet Med Anim Health. 2016;8(4):29–34. doi:10.5897/JVMAH2014-0315
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
