Back to Journals » Infection and Drug Resistance » Volume 16
Antibacterial Activity of Thesium chinense Turcz Extract Against Bacteria Associated with Upper Respiratory Tract Infections
Authors Wei J, Zhang C , Ma W, Ma J, Liu Z, Ren F, Li N
Received 11 June 2023
Accepted for publication 2 August 2023
Published 8 August 2023 Volume 2023:16 Pages 5091—5105
DOI https://doi.org/10.2147/IDR.S425398
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Juanru Wei,1,* Cong Zhang,2,* Wei Ma,1 Juncheng Ma,1 Zhenzhen Liu,1 Fucai Ren,1 Ning Li1
1School of Pharmacy, Anhui Medical University, Hefei, 230032, People’s Republic of China; 2School of Basic Medical Sciences, Anhui Medical University, Hefei, 230032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ning Li; Fucai Ren, School of Pharmacy, Anhui Medical University, No. 81 Meishan Road Shushan District, Hefei, 230032, People’s Republic of China, Tel/Fax +86-5516-516-1115, +86-5516-517-2133, Email [email protected]; [email protected]
Purpose: The drug resistance of Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes and Haemophilus influenzae has become more and more serious, and it is urgent to seek new antibacterial drugs. In this study, Thesium chinense Turcz. extracts were tested for its potential antibacterial activities.
Methods: T. chinense powder was extracted with 5 solvents of different polarity (ethyl alcohol, petroleum ether, ethyl acetate, n-butyl alcohol and double distilled water), and their antibacterial activities were tested. The Broth dilution method was used to evaluate the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of highly active plant extracts with a concentration of 1g/mL. The inhibitory activity of this extract on biofilm formation was investigated. Afterwards, we investigated its effect on the transcriptome of S. aureus.
Results: The ethanol extract coded as BRY, only inhibited S. aureus, whereas the ethyl acetate extract coded as BY2 showed inhibitory effect on all the tested bacteria. The MIC of BRY on S. aureus was 128 mg/mL, and the MBC was 512 mg/mL. The MIC of BY2 against S. aureus, S. pneumoniae, S. pyogenes and H. influenzae were 8 mg/mL, 4 mg/mL, 4 mg/mL, and 4 mg/mL, respectively. The MBC of BY2 for these four bacteria ranged from 4 to 256 mg/mL. Mechanism studies have shown that BRY and BY2 have an impact on anti-formation of biofilms at MIC concentrations. Transcriptome sequencing results showed that 531 genes were up-regulated and 340 genes showed down-regulated expression in S. aureus after BY2 treatment.
Conclusion: BY2 has a broader antibacterial spectrum than BRY. Meanwhile, the inhibitory effect of BY2 on S. aureus is better than BRY. The mechanism of BY2 against S. aureus may relate to its inhibition of ribosome synthesis, restriction of key enzymes of citric acid cycle, decrease of pathogenicity and influence on biofilm formation. The results confirmed that BY2 was the main antibacterial part of T. chinense, which can be used as a source of antibacterial agents.
Keywords: plant extract, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae, antibacterial
Graphical Abstract:
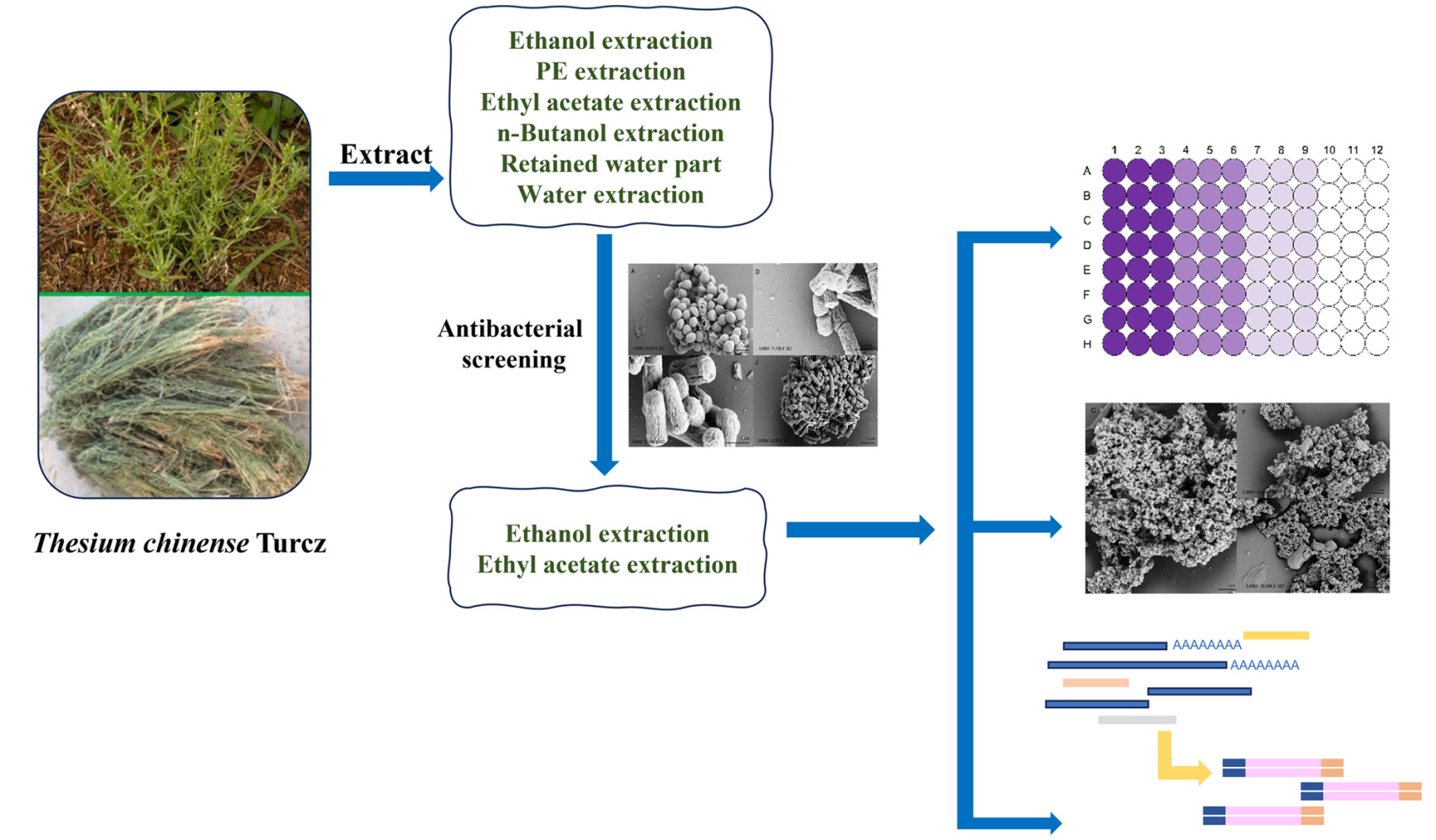
Introduction
Upper respiratory tract infections (URTIs) are primarily caused by bacterial and viral pathogens, such as Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, Pseudomonas aeruginosa, Klebsiella pneumoniae, Streptococcus pyogenes, influenza virus (IFV), and respiratory syncytial virus (RSV).1–3 When infected with these pathogens, people may develop sinusitis, pharyngitis, tonsillitis, or otitis media.4,5 Among the pathogens that cause UTRIs, bacteria is one of the most threatening to human health. A study of deaths from bacterial pathogen infections found that S. aureus infections were the leading cause of death from bacterial infections in 135 countries, and the deadliest bacteria in people over the age of 15 worldwide. Similarly, S. pneumoniae is the pathogen associated with the majority of deaths in children younger than 5 years of age.6 In addition to infecting the respiratory tract alone, bacterial pathogens can cause secondary infections following viral infection. About one-third of viral respiratory infections were caused by influenza viruses.1 After influenza virus infection, approximately 50 to 90% of severe or fatal influenza infections were associated with bacterial co-infection or secondary infection.7–9 At least 15 bacterial strains have been reported to be associated with viral respiratory tract infections. The main bacterial strains that increased the risk of death from viral infection were S. aureus, S. pneumoniae, S. pyogenes, and H. influenzae.10–14
It should be emphasized that antimicrobial resistance is rapidly emerging and spreading globally. There have been numerous reports of resistance in S. aureus, S. pneumoniae, S. pyogenes, and H. influenzae.15–18 As a result, we need to develop new antimicrobial drugs. In previous studies, medicinal plant extracts have shown good antibacterial effects, which may offer some solutions to the drug resistance problem.19,20
China has a long history of using botanicals for antibacterial treatment. Thesium chinense Turcz. is a traditional Chinese medicine plant in the genus Thesium of family Santalaceae. It is a perennial herb and is commonly used to treat URTIs in traditional Chinese medicine (TCM),21,22 which is also known as a “natural antibiotic”.23 T. chinense was first recorded about a thousand years ago in a TCM monograph called Bencao Tujing. According to Bencao Tujing, T. chinense promotes lactation and regulates bodily functions. This is the embodiment of the ancient use of T. chinense for acute mastitis. Bacterial infection is an important cause of acute mastitis. Most patients with acute mastitis are found to have pathogenic bacteria in their milk or pus.24 Besides, according to Guoyao Tiyao, it can be used to treat cervical lymphadenitis which is often caused by infection with S. pyogenes or S. aureus.25 In addition, phytochemical studies in recent years have shown that (12E) -heptadec-12-en-8,10-diynoic acid, dodec-9,11-diynoic acid and exocarpic acid in T. chinense have inhibitory effects on oral bacterial infection.26 Aqueous extracts of T. chinense have also been shown to have antibacterial activity against some Gram-positive and Gram-negative bacteria. Similarly, Yuan et al27 reported that water extracts of T. chinense showed inhibitory effects on Escherichia coli, S. aureus, and Bacillus thuringiensis. Therefore, T. chinense has a good antibacterial effect.28
In this paper, we studied the antibacterial effect of six extracts from T. chinense (ethanol, petroleum ether, ethyl acetate, n-butanol and double distilled water) against four bacteria closely related to respiratory tract infection (Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes and Haemophilus influenzae), in order to explore new natural drugs against respiratory tract infection-related bacteria and solve the urgent problem of antibiotic resistance. We hope that this study can provide new therapeutic ideas for the clinic.
Materials and Methods
Plant Materials
The whole dry herb of T. chinense was collected from Xiangyang city, Hubei Province, P. R. China in May 2019. The species was identified by Prof. Kai-Jin Wang of Anhui University, and a voucher specimen (No. 20190927) was deposited in the School of Pharmacy, Anhui Medical University.
Ethanol Extract Preparation
Plant materials (10 kg) were sequentially soaked in ethanol (95%, 85%, 75%, 1:10 w/v) at room temperature for 12 h, and ultrasonically extracted at 60°C for 4 h. The filtrate was combined, filtered, and evaporated with a rotary evaporator. The extract was named BRY.
Preparation of Other Extracts
A portion of BRY (23 kg) was suspended with 23L water, and then sequentially extracted 4 times (1:1 v/v, 30 min) with petroleum ether (PE), ethyl acetate (EA) and n-butanol. The organic phases were evaporated and freeze-dried to obtain BS1, BY2 and BZ3, respectively. The last remaining aqueous phase was collected and lyophilized and named as BS4.
The filter residue in “Ethanol extract preparation” was extracted with double distilled water (1:1 w/v, 4 h) at 60°C for 3 times. The filtrate was collected, and finally freeze-dried, named as BRS.
The prepared samples were dissolved in sterile PBS and filtered at 0.22 μm.
Bacterial Strains and Culture Conditions
S. aureus (ATCC 29213) was obtained from the First Affiliated Hospital of Anhui Medical University. S. pneumoniae (ATCC 49619), S. pyogenes (ATCC 19615) and H. influenzae (ATCC 49766) were purchased from Hefei MORE Bio.
S. aureus was cultured in Mueller–Hinton broth (MHB). S. pneumoniae, S. pyogenes and H. influenzae were cultured in Mueller–Hinton broth (MHFB) supplemented with 5% lysed horse blood and 20 mg/L β-NAD.29 The strains were cultured at 37°C, 200 rpm.
Preliminary Screening of Antibacterial Activity
Bacterial suspension (106 CFU/mL) was prepared with sterile medium. An equal volume of bacterial suspension was added to agar medium and allowed to dry. Wells were made with a sterile borer in the inoculated agar plates. Extracts (100 μL) were propelled into the wells for each test plates.30 The plates were incubated at 37°C, 5% CO2 for 24 h, and then the diameter of the inhibition zone was measured with a vernier caliper.
Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
MIC was measured by broth microdilution31 in 96-well microtiter plates. A 100 μL of each bacterial suspension (2 × 106 CFU/mL) was incubated 24 h with 100 μL aliquot of test concentration of each extract prepared before.
Inoculum (100 μL) from wells without turbidity was spread onto the agar plate at 37 °C for 24 h. The number of the colony-forming units was counted. There will be no colony formation on the plate of the MBC.32
Determination of Time–Kill Curves
The 96-well microtiter plates were containing bacterial culture (1 × 106 CFU/mL, final concentration) and 100 μL extract prepared before. The final concentration of extract was 0.5 × MIC and 1 × MIC, respectively. The bacteria-containing PBS without extract was set as the negative control group, and PBS was used to be solvent control. Then, the absorbance value was measured at 630 nm (Synergy™ HTX) at different time points (0, 2, 4, 8, 12 and 24 h), and a growth curve was drawn.
Biofilm Formation Inhibition Assay
Bacteria with or without extracts were cultured in 96-well plates at 37°C for 24 h. The supernatant was discarded to remove free bacteria. Bacteria were fixed with methanol for 20 min, stained with crystal violet (0.1%), washed with PBS after 20 min to remove unbound crystal violet solution. Finally, absolute ethanol was added to the 96-well plate. Plates were incubated at 37°C and 200 rpm for 30 minutes. The absorbance value was measured at 590 nm.
Determination of CFU of Bacteria in Biofilms
The treatment of biofilms was as described in “Biofilm formation inhibition assay”. Free bacteria were discarded gently and 200 μL PBS was used to resuspend the bacteria on the biofilm. The bacterial suspension was collected and inoculated on agar plates. CFU was calculated after 24 h.
MTT Metabolism Detection
After treating the biofilm as described in “Biofilm formation inhibition assay”, the free bacteria were discarded. A 20 μL MTT (5 mg/mL) was added to each well, and the plates were incubated in the dark for 4 h. After that, MTT was discarded, and DMSO was added. Plates were incubated at 37°C for 15 min. The absorbance value was measured at 570 nm.
SEM
The 0.5 × MIC extracts were co-cultured with bacterial suspensions with a final concentration of 106 CFU/mL. Bacterial suspension without drug was taken as negative control. The bacterial strains were cultivated at 37°C and 200 rpm for 24 h. After centrifugation, the bacteria were immobilized with 2.5% glutaraldehyde at 4°C for 8–12 h. The sample was dehydrated with different concentrations of ethanol (10%, 30%, 50%, 70%, 80%, 90%, 100%) and then dehydrated with HMDS. After coated with gold-palladium, the sample was observed and photographed with a scanning electron microscope (ZEISS Gemini SEM 300, Germany).
Pretreatment of Transcriptome Sequencing Samples
Transcriptome sequencing was performed on S. aureus treated with BY2.
The bacterial suspension was adjusted by 0.5 McIntosh units. The drug was added to a prepared bacterial suspension with a bacterial concentration of 106 CFU/mL and a final drug concentration of 0.5 × MIC. The bacterial suspension without drug was taken as the negative control. The suspensions were cultured at 37°C and 200 rpm for 24 h.
After centrifugation at low temperature, the supernatant was discarded, and the sample was cleaned with PBS 3 times to remove the medium. The sample was quickly frozen with liquid nitrogen at low temperature and stored in the refrigerator at −80°C. For each group, 3 biological samples were prepared for repeated testing. The above samples were transferred on dry ice to the high-throughput laboratory of Wuhan BGI Technology Service Co., LTD for transcriptome sequencing and follow-up analysis.
Transcriptome Sequencing and Analysis Table
The clean reads were compared to the reference genome for the mapped data. Then, quality assessments were performed, and differential expression genes (DEGs) were detected between groups, for bioinformatics analyses of functional annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways and functional enrichment.
Real-Time PCR Assay for Gene Expression Analysis
The bacteria were cultured for 24 hours according to the method described in “Pretreatment of transcriptome sequencing samples”. After centrifugation, the total RNA of S. aureus was isolated using an RNA extraction kit as described. Reverse transcription of total RNA into cDNA.33 RT-qPCR was used to detect gene expression. 16S RNA was used as housekeeper gene, and the results were expressed by 2−ΔΔCt. All primer sequences are shown in Table 1.
 |
Table 1 List of Primer Sequences Used for Quantitative Real-Time PCR (qRT-PCR) |
Statistical Analysis
All data were analyzed using GraphPad Prism 8. Significant differences were calculated using one-way ANOVA, and P < 0.05 was considered statistically significant.
Results
Antimicrobial Activity
The diameters of the inhibition zones of BRY, BS1, BY2, BZ3, BS4 and BRS against the four bacteria are shown in Table 2. BRY and BY2 have shown antibacterial activity against S. aureus. Moreover, BY2 also has an inhibitory effect on S. pneumoniae, S. pyogenes, and H. influenzae. No significant inhibitory zone was detected for other extracts. Therefore, BRY and BY2 were considered to be highly active extracts, and they were used to conduct subsequent studies.
 |
Table 2 The Diameter of the Inhibition Zone |
The MICs and MBCs of BRY and BY2 against the four strains were determined by the broth microdilution method (Table 3, Figure 1), the MIC of BRY against S. aureus was 128 mg/mL, and the MBC was 512 mg/mL. BY2 inhibited the growth of S. aureus, S. pneumoniae, S. pyogenes, and H. influenzae at MICs of 8 mg/mL, 4 mg/mL, 4 mg/mL, and 4 mg/mL, respectively. The MBC values were 16 mg/mL, 4 mg/mL, 256 mg/mL, and 64 mg/mL, respectively.
 |
Table 3 Antibacterial Activities of BRY and BY2 |
 |
Figure 1 The growth of bacteria strains on solid medium (BRY and BY2 at MICs and MBCs). Notes: (A–F) S. aureus, (G–I) S. pneumoniae, (J–L) S. pyogenes, (M–O) H. influenzae. |
Time–Killing Curves
The time–killing curves for BRY and BY2 against bacteria are shown in Figure 2. At a BRY concentration of 0.5 × MIC, S. aureus grows slowly in the range 0–12 h. However, bacterial growth became faster after 12 h, and the OD630nm value at 24 h was somewhat similar with the negative control. As the concentration is increased to 1 × MIC, the OD630nm value of the bacterial solution is very low, indicating that very few bacteria grow. Unlike BRY, BY2 also shows a good inhibitory effect on S. aureus at 0.5 × MIC concentration.
 |
Figure 2 Time–killing curves for BRY and BY2 against tested bacteria at 1 × MIC and 0.5 × MIC. Notes: (A and B) S. aureus, (C) S. pneumoniae, (D) S. pyogenes, (E) H. influenzae. |
The OD630nm values for S. pneumoniae, S. pyogenes, and H. influenzae demonstrate that bacterial growth is still significantly suppressed when the BY2 concentration is at 1 × MIC. However, when the concentration is 0.5 × MIC, the growth of the bacteria accelerates at 8h, and eventually the results are similar to those of the negative control.
Bacterial Biofilm Inhibition Results
Bacterial pathogenicity is associated with a number of factors, including biofilm formation. Therefore, we performed a biofilm formation disruption experiment. As shown in Figure 3, BRY and BY2 appear to have disrupted the formation of bacterial biofilms at MICs. However, the effect was less pronounced at a concentration of 0.5 × MIC.
 |
Figure 3 BRY and BY2 inhibits the biofilm formation of tested bacteria. Notes: (A) S. aureus, (B) S. pneumoniae, (C) S. pyogenes, (D) H. influenzae. **Compared with “Negative Control”, P < 0.01. |
According to the inhibitory effect of BRY and BY2 on bacterial biofilm formation, we further studied the effect of BRY and BY2 on the bacterial viability in biofilm (Figure 4) and the growth of bacteria on solid medium after treatment (Figure 5). BRY and BY2 (1 × MIC) significantly reduced the bacterial activity in biofilm of S. aureus, and the effect of BY2 was better than that of BRY. BY2 performed similarly on the other three tested bacteria, with a significant reduction in cell viability at 0.5 × MIC and 1 × MIC.
 |
Figure 5 CFU of bacteria biofilm. Notes: (A) S. aureus, (B) S. pneumoniae, (C) S. pyogenes, (D) H. influenzae. *Compared with “Negative Control”, P<0.05. **Compared with “Negative Control”, P<0.01. |
Both BRY and BY2 were able to reduce the number of bacteria in the biofilm of the tested bacteria at 1 × MIC. BY2 (0.5 × MIC) is also effective in reducing bacteria in biofilms of S. pyogenes and H. influenzae.
Observation of Bacteria
SEM results (Figure 6) showed that the bacteria without drug treatment (negative control) grew well. Bacteria were severely damaged after BRY or BY2 treatment (1 × MIC). The antimicrobial effect of BRY and BY2 can be evaluated by the fact that only a very small number of bacteria survive.
 |
Figure 6 SEM visualization of bacteria. Notes: (A–C) S. aureus, (D–F) S. pneumoniae, (G–I) S. pyogenes, (J–L) H. influenzae. |
Analysis of Transcriptome Sequencing Results
RNA sequencing was used to analyze the transcriptome of S. aureus. The differentially expressed genes between the control group and the experimental group were analyzed. The results showed that a total of 1019 genes were changed in S. aureus treated with BY2 compared with the untreated sample control, of which 531 genes were up-regulated and 340 genes showed down-regulated expression (Q value < 0.05 and log2-fold-change > 1 or < −1). The volcano plot of differential genes is shown below (Figure 7), with upregulation in red and downregulation in blue. In order to verify the accuracy of the results of RNA-Seq, some differentially expressed genes were selected randomly for RT-qPCR, and 16S RNA was used as the housekeeping gene. The results of RT-qPCR were consistent with the trend of gene expression by RNA-seq (Figure 8). To analyze the antibacterial mechanism of BY2 against S. aureus, gene ontology (GO) analysis (Figure 9A) and KEGG analysis (Figure 9B) were performed, respectively. GO results showed that after BY2 treating, DEGs were mainly enriched in cellular process, metabolic process, localization and biological regulation. They contained 641, 580, 120 and 114 genes, respectively. The up-regulated genes involved 121 pathways, which were mainly enriched in 15 pathways, of which 3 pathways were significantly enriched (Q-value < 0.05), namely photosynthesis (8 genes), amino acid biosynthesis (53 genes) and ribosome (33 genes). Down-regulated genes were mainly involved in 131 pathways, and the main enrichment pathways included citric acid cycle (15 genes), carbon metabolism (40 genes) and life span regulation (6 genes) (p < 0.01), including 641, 580, 120 and 114 genes, respectively.
Discussion
T. chinense extract is believed to have a good inhibitory effect on Gram-positive bacteria. Additionally, it is sensitive to Aeromonas hydrophila, a gram-negative bacterium.28 Another study reported that it showed antimicrobial activity against the proliferation of three oral pathogens, including Porphyromonas gingivalis, Fusobacterium nucleatum and Streptococcus mutans.26 This suggests that T. chinense have a broad-spectrum antibacterial effect. Therefore, we conjecture that it also has an inhibitory effect on other bacteria.
In this paper, we selected several bacteria that caused the most deaths due to co-infection or secondary infection in the influenza pandemic, which were S. aureus, S. pneumoniae, S. pyogenes and H. influenzae, and evaluated the antibacterial activity of each extract part of T. chinense. Different from the previous studies where only one or two solvents were used for extraction,34 this time, ethanol extraction was performed first. The filtered and filtered residuals of the ethanol extract were then extracted using PE, EA, n-butanol and ddH2O, respectively. The final water section was retained. Eventually, six different fractions of the extract were obtained.
Clearly, the results of in vitro experiments using the punch method to measure the inhibitory zone suggested that BRY has an inhibitory effect on S. aureus, in agreement with the results of Liu et al.27 However, the diameter of the suppression zone of BRY was not detected for other strains. The antibacterial zone diameters of BY2 against S. aureus, S. pneumoniae, S. pyogenes and H. influenzae were 13.27±0.68mm, 16.4±1.78mm, 16.57±0.54mm and 17.23±0.34mm, respectively. The inhibition of BS1, BZ3, BS4, and BRS were not available. Therefore, the next experiments were performed only on BRY and BY2. The effect of BRY and BY2 on bacterial growth could be visually observed through dynamic time–kill curves. Especially, when the concentration was 1 × MIC, the absorbance results indicated that there is almost no bacterial growth. On this basis, crystal violet staining detection (Figure 3) and scanning electron microscope observation (Figure 6) were carried out. Crystal violet is a basic dye that binds to nucleic acids in nuclei and stains them blue. Based on the results of crystal violet staining, it can be determined that BRY disrupts the biofilm formation of S. aureus. The biofilm destruction effect was strong for a concentration of 1 × MIC, which was also verified by the SEM results. Compared to BRY, BY2 disrupts the biofilm formation of more bacterial strains.
Biofilms are fixed microbial communities attached to a surface. In the respiratory tract, bacteria localize in communities of biofilms.35 Biofilm formation is one of the main epidemiological factors in bacterial infection. The presence of biofilms can impede the entry of some immune defenses, for example, causing macrophages to incompletely penetrate the biofilm matrix and appear to be “frustrated phagocytosis”. Bacteria in biofilms are more resistant to current treatments than planktonic bacteria. In addition, biofilm formation has the potential to increase patient mortality. Severe infection with methicillin-resistant S. aureus has a mortality rate of approximately 20%–25%.36–40
BRY and BY2 not only disrupted bacterial biofilm formation, but also reduced bacterial viability in the biofilm (Figure 5). In other words, after treatment with two extracts, the number of bacteria in the biofilm decreased significantly. It means that T. chinense may have the potential to alleviate the pressure of antibiotic resistance.
By analyzing the transcriptome sequencing results of S. aureus treated and untreated with BY2, combined with GO functional annotation analysis and KEGG pathway enrichment analysis, we found significant enrichment of up-regulated genes in photosynthesis, amino acid biosynthesis, and ribosome. The photosynthetic pathway is mainly associated with the F-type H+/Na+ ATPase. The F-type ATPase gene is found primarily in the plasma membranes of plants and bacteria. It is powered by the ATP. A large number of genes were enriched in amino acid biosynthesis pathways, mostly in glycolysis and amino acid metabolism, such as gap, ilvd, leuB, aroK, trpD, etc. The expression of glyceraldehyde-3-phosphate dehydrogenase was up-regulated, which facilitates the conversion of three carbon compounds from glyceraldehyde-3p to pyruvate. At the same time, the expression of 3-isopropyl malate dehydrogenase is increased, which promotes the production of branched-chain amino acids. Genes enriched in the ribosomal pathway included rplE and rpsL, which encode ribosomal component proteins. The ribosome is an important organelle for bacteria to survive and consists of three rRNA chains, 16S, 23S and 5S. 23S rRNA, 5S rRNA, and more than 30 proteins together form the large 50S subunit, and the small 30S subunit and large 50S subunit combine to form the 70S complex. Ribosome is known as the “molecular machine” of protein synthesis, which converts the genetic code into amino acid sequences and constructs protein polymers from amino acid monomers, which is of great significance to cell life activities.41 In this study, BY2 could cause a significant up-regulation of ribosome-related genes after acting on S. aureus (Figure 7B), which affected the process of bacterial ribosomal protein synthesis. In the enrichment analysis of down-regulated genes, we did not find significantly enriched pathways (Qvalue > 0.05). The analysis results indicate that the number of genes annotated as part of the KEGG pathway in this strain is relatively small, which may contribute to the distortion of statistical significance. Therefore, pathways with P < 0.01 were analyzed here. The down-regulated enrichment results for the differentially expressed genes indicate that the pathways primarily involved include citric acid cycle, carbon metabolism, and lifespan regulation. The citric acid cycle is an important aerobic pathway that is the final step in the metabolism of sugar, fat, and amino acids.42 In a series of reactions, the two carbons in citrate are oxidized to carbon dioxide, and this reaction pathway provides NADH for oxidative phosphorylation and other metabolic processes. After treating S. aureus with BY2, the expression of 15 genes involved in the citric acid cycle was down-regulated. In particular, the expression of citrate synthase, isocitrate dehydrogenase, dihydrolipoamide succinyltransferase and dihydrolipoamide dehydrogenase, which are involved in the formation of the pyruvate dehydrogenase complex, were significantly down-regulated. Furthermore, we note that the expressions for clpP and dnaK are similarly suppressed. ClpP is one of the housekeeping genes regulating the virulence of S. aureus.43 By inhibiting the expression of the clpP gene, bacterial pathogenicity can be reduced and thus antibacterial effects can be enhanced. In addition, it has been pointed out that clpP and its molecular chaperone dnaK can affect bacterial biofilm formation.44 Therefore, we suggest that BY2 shows good antimicrobial activity against S. aureus, which may be related to its effect on ribosome synthesis, limiting key enzymes of the citric acid cycle, reducing pathogenicity and affecting biofilm formation.
However, the lack of in vivo experiments and the lack of further identification and characterization of the active compounds are limitations of this study. We will also continue to conduct research to provide insights into the next step.
Conclusion
This study demonstrated the antibacterial activity of T. chinense extracts against S. aureus, S. pneumoniae, S. pyogenes and H. influenzae. The results showed that ethanol extract and ethyl acetate extract inhibited in vitro growth of at least one or more of the tested bacteria. Ethyl acetate extract effectively inhibited the growth of four bacterial species, indicating that the metabolites in the extract have broad spectrum of antibacterial activity. It had low MIC values for all the tested bacteria. In addition, the growth of S. aureus was inhibited when ethyl acetate extract was at 0.5 × MIC. The results of RNA-seq indicate that the inhibitory mechanism of ethyl acetate extract on S. aureus growth may be related to its inhibition of ribosomal synthesis, restriction of key enzymes in the citric acid cycle, reduction of pathogenicity, and influence on biofilm formation. The overall results substantiate the use of T. chinense as an antibacterial agent. After a more in-depth analysis of isolated and characterized active compounds, this traditional plant could be the best candidate for further research in the development of new antimicrobial agents.
Abbreviations
BMD, broth microdilution; BRS, water extract of Thesium Chinense Turcz; BRY, ethanol extract of T. chinense; BS1, petroleum ether extract of T. chinense; BS4, the remaining aqueous phases of BZ3; BY2, ethyl acetate extract of T. chinense; BZ3, n-butanol extract of T. chinense; ddH2O, double distilled water; EA, ethyl acetate; g, gramme; HMDS, Hexamethyldisilazane; MBC, minimum bactericidal concentration; mg, milligramme; MHB, Mueller–Hinton broth; MHFB, Mueller–Hinton broth supplemented with lysed horse blood and β-NAD; MIC, minimum inhibitory concentration; MTT, 3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl Tetrazolium Bromide; NA, not applicable; PBS, phosphate-buffered saline; PE, petroleum ether; SEM, scanning electron microscopy; URTIs, upper respiratory tract infections; β-NAD, β-Nicotinamide adenine dinucleotide.
Ethics Approval and Informed Consent
Research on plants, including the collection of plant material, is conducted in accordance with guidelines provided by the author’s institution and national or international regulations.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was supported by the National Science Foundation of China (No. 32270424), Major Project of Science and Technology of Anhui Province, China (202203a07020014), and Scientific Research Platform Improvement Project of Anhui Medical University (2022xkjT045).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Li ZJ, Zhang HY, Ren LL, et al. Etiological and epidemiological features of acute respiratory infections in China. Nat Commun. 2021;12(1):5026. doi:10.1038/s41467-021-25120-6
2. Zhang N, Wang L, Deng X, et al. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92(4):408–417. doi:10.1002/jmv.25674
3. Lu M. Etiology of bacterial pneumonia in children. Chin J Pract Pediatr. 2018;33(09):702–706. doi:10.19538/j.ek2018090611
4. Jain N, Lodha R, Kabra SK. Upper respiratory tract infections. Indian J Pediatr. 2001;68(12):1135–1138. doi:10.1007/BF02722930
5. Bakaletz LO. Viral-bacterial co-infections in the respiratory tract. Curr Opin Microbiol. 2017;35:30–35. doi:10.1016/j.mib.2016.11.003
6. GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2023;400(10369):2221–2248. doi:10.1016/S0140-6736(22)02185-7
7. Wu P, Presanis AM, Bond HS, Lau EHY, Fang VJ, Cowling BJ. A joint analysis of influenza-associated hospitalizations and mortality in Hong Kong, 1998-2013. Sci Rep. 2017;7(1):929. doi:10.1038/s41598-017-01021-x
8. McDanel JS, Perencevich EN, Storm J, et al. Increased Mortality Rates Associated with Staphylococcus aureus and Influenza Co-infection, Maryland and Iowa, USA(1). Emerg Infect Dis. 2016;22(7):1253–1256. doi:10.3201/eid2207.151319
9. McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12(4):252–262. doi:10.1038/nrmicro3231
10. Madhi SA, Schoub B, Klugman KP. Interaction between influenza virus and Streptococcus pneumoniae in severe pneumonia. Expert Rev Respir Med. 2008;2(5):663–672. doi:10.1586/17476348.2.5.663
11. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi:10.1086/591708
12. Jamieson AM, Pasman L, Yu S, et al. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340(6137):1230–1234. doi:10.1126/science.1233632
13. Klein EY, Monteforte B, Gupta A, et al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10(5):394–403. doi:10.1111/irv.12398
14. Borgogna TR, Hisey B, Heitmann E, Obar JJ, Meissner N, Voyich JM. Secondary Bacterial Pneumonia by Staphylococcus aureus Following Influenza A Infection Is SaeR/S Dependent. J Infect Dis. 2018;218(5):809–813. doi:10.1093/infdis/jiy210
15. Abbara S, Guillemot D, El Oualydy S, et al. Antimicrobial Resistance and Mortality in Hospitalized Patients with Bacteremia in the Greater Paris Area from 2016 to 2019. Clin Epidemiol. 2022;14:1547–1560. doi:10.2147/CLEP.S385555
16. Ahmed N, Khan M, Saleem W, et al. Evaluation of Bi-Lateral Co-Infections and Antibiotic Resistance Rates among COVID-19 Patients. Antibiotics. 2022;11(2):276. doi:10.3390/antibiotics11020276
17. Zhou Y, Wang Y, Cheng J, Zhao X, Liang Y, Wu J. Molecular epidemiology and antimicrobial resistance of Haemophilus influenzae in Guiyang, Guizhou, China. Front Public Health. 2022;10:947051. doi:10.3389/fpubh.2022.947051
18. Hascelik G, Soyletir G, Gulay Z, et al. Serotype distribution of Streptococcus pneumoniae and pneumococcal vaccine coverage in adults in Turkey between 2015 and 2018. Ann Med. 2023;55(1):266–275. doi:10.1080/07853890.2022.2160877
19. Cowan MM. Plant Products as Antimicrobial Agents. Clin Microbiol Rev. 1999;12(4):564–582. doi:10.1128/CMR.12.4.564
20. Lee JH, Kim YG, Khadke SK, Yamano A, Woo JT, Lee J. Antimicrobial and antibiofilm activities of prenylated flavanones from Macaranga tanarius. Phytomedicine. 2019;63:153033. doi:10.1016/j.phymed.2019.153033
21. Editorial Board of Chinese Materia Medica. Chinese Materia Medica. Shanghai Science and Technology Press; 1999.
22. Flora of China Editorial Committee. Flora of China. Science Press. 80. 1988.
23. Chen P, Chen X, Wu C, Meng Y, Cao J. Research progress on the development and utilization of Thesium chinense Turcz. Chin Wild Plant Resour. 2020;39(06):48–52.
24. Li D, Li J, Yuan Y, et al. Risk factors and prognosis of acute lactation mastitis developing into a breast abscess: a retrospective longitudinal study in China. PLoS One. 2022;17(9):e0273967. doi:10.1371/journal.pone.0273967
25. Leung AK, Davies HD. Cervical lymphadenitis: etiology, diagnosis, and management. Curr Infect Dis Rep. 2009;11(3):183–189. doi:10.1007/s11908-009-0028-0
26. Liu C, Li XT, Cheng RR, et al. Anti-oral common pathogenic bacterial active acetylenic acids from Thesium chinense Turcz. J Nat Med. 2018;72(2):433–438. doi:10.1007/s11418-018-1180-3
27. Yuan Y, Long Z, Xu X, Wang L, Ying M. Comparison of wild and cultured thesium chinense turcz on bacteriostasis and anti-inflammation. Chin J Pharm Biotec. 2006;3:219–222. doi:10.19526/j.cnki.1005-8915.2006.03.015
28. Liu Y, Pan L, Qi K, Jiang L. Sensitivity test of effective extracts from Thesium chinense to seven kinds of bacteria. Guizhou Med. 2006;6:564–566. doi:10.3969/j.issn.1000-744X.2006.06.043
29. The European Committee on Antimicrobial Susceptibility Testing. Media preparation for EUCAST disk diffusion testing and for determination of MIC values by the broth microdilution method. Available from: https://www.eucast.org/ast_of_bacteria/media_preparation.
30. Besra M, Kumar V. In vitro investigation of antimicrobial activities of ethnomedicinal plants against dental caries pathogens. 3 Biotech. 2018;8(5):257. doi:10.1007/s13205-018-1283-2
31. The European Committee on Antimicrobial Susceptibility Testing. EUCAST reading guide for broth microdilution. Available from: https://www.eucast.org/ast_of_bacteria/media_preparation.
32. Rodriguez-Melcon C, Alonso-Calleja C, Garcia-Fernandez C, Carballo J, Capita R. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) for Twelve Antimicrobials (Biocides and Antibiotics) in Eight Strains of Listeria monocytogenes. Biology. 2021;11(1):46. doi:10.3390/biology11010046
33. Mutonga DM, Mureithi MW, Ngugi NN, Otieno FCF. Bacterial isolation and antibiotic susceptibility from diabetic foot ulcers in Kenya using microbiological tests and comparison with RT-PCR in detection of S. aureus and MRSA. BMC Res Notes. 2019;12(1):244. doi:10.1186/s13104-019-4278-0
34. Parveen Z, Deng Y, Saeed MK, Dai R, Ahamad W, Yu YH. Antiinflammatory and analgesic activities of Thesium chinense Turcz extracts and its major flavonoids, kaempferol and kaempferol-3-O-glucoside. Yakugaku Zasshi. 2007;127(8):1275–1279. doi:10.1248/yakushi.127.1275
35. Welp AL, Bomberger JM. Bacterial Community Interactions During Chronic Respiratory Disease. Front Cell Infect Microbiol. 2020;10:213. doi:10.3389/fcimb.2020.00213
36. Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi:10.1146/annurev-med-042711-140023
37. Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22(6):326–333. doi:10.1016/j.tim.2014.02.001
38. Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7(4):493–512. doi:10.4155/fmc.15.6
39. Vaughn AR, Haas KN, Burney W, et al. Potential Role of Curcumin Against Biofilm-Producing Organisms on the Skin: a Review. Phytother Res. 2017;31(12):1807–1816. doi:10.1002/ptr.5912
40. Mouwakeh A, Kincses A, Nove M, et al. Nigella sativa essential oil and its bioactive compounds as resistance modifiers against Staphylococcus aureus. Phytother Res. 2019;33(4):1010–1018. doi:10.1002/ptr.6294
41. Zhang L, He J, Bai L, Ruan S, Yang T, Luo Y. Ribosome-targeting antibacterial agents: advances, challenges, and opportunities. Med Res Rev. 2021;41(4):1855–1889. doi:10.1002/med.21780
42. Wang L, Qu F, Zhu Z, et al. The important role of tricarboxylic acid cycle metabolism pathways and core bacterial communities in carbon sequestration during chicken manure composting. Waste Manag. 2022;150:20–29. doi:10.1016/j.wasman.2022.06.034
43. Michel A, Agerer F, Hauck CR, et al. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol. 2006;188(16):5783–5796. doi:10.1128/JB.00074-06
44. Pu Y, Li Y, Jin X, et al. ATP-Dependent Dynamic Protein Aggregation Regulates Bacterial Dormancy Depth Critical for Antibiotic Tolerance. Mol Cell. 2019;73(1):143–156 e4. doi:10.1016/j.molcel.2018.10.022
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




