Back to Journals » International Journal of General Medicine » Volume 14
Analysis of Communal Molecular Mechanism and Potential Therapeutic Targets in Heart Failure and Type 2 Diabetes Mellitus
Authors Wang Z, Gu Y, Sun Y, Xu Y , Zhang M, Jiang T
Received 25 June 2021
Accepted for publication 29 September 2021
Published 9 October 2021 Volume 2021:14 Pages 6549—6561
DOI https://doi.org/10.2147/IJGM.S325339
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Zuoxiang Wang,1,2 Yiyu Gu,1,2 Yunjuan Sun,1 Yinan Xu,1,2 Mingyang Zhang,1,2 Tingbo Jiang1
1Department of Cardiology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China; 2Department of Medicine, Soochow University, Suzhou, Jiangsu, People’s Republic of China
Correspondence: Tingbo Jiang
Department of Cardiology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China
Tel +86 18906201122
Email [email protected]
Background: Although increasing evidence has suggested an interaction between heart failure (HF) and Type 2 diabetes mellitus (T2DM), the common mechanisms of the two diseases remain unclear. Therefore, this study aimed to obtain the differentially expressed genes (DEGs) and potential biomarkers or therapeutic targets in HF and T2DM.
Methods: The communal DEGs of HF and T2DM were identified by analyzing the two microarray datasets (GSE84796 and GSE95849), and functional annotation was performed for the communal DEGs to uncover the potential molecular mechanisms of HF and T2DM. Subsequently, STRING database and Cytoscape software were used to construct the protein–protein interaction (PPI) network and screen the hub genes. Finally, co-expression and drug–gene interaction prediction analysis and mRNA–miRNA regulatory network analysis were performed for hub genes.
Results: A total of 233 up-regulated genes and 3 down-regulated genes were found between HF and T2DM. The functional enrichment of DEGs and genes in each four modules were mainly involved in immunity. In addition, five hub genes were identified from PPI network, including SYK, SELL, RAC2, TLR8 and ITGAX.
Conclusion: The communal DEGs and hub genes identified in this research contribute to discover the underlying biological mechanisms and presents potential biomarkers or therapeutic targets in HF and T2DM.
Keywords: bioinformatical analysis, therapeutic targets, heart failure, type 2 diabetes mellitus
Introduction
Heart failure (HF) is the inability of the heart to transport sufficient amounts of blood to meet the demands of our body at normal filling pressures, finally causing a complex and severe disease syndrome.1,2 HF is a rapidly growing public health issue with an estimated prevalence of >37.7 million individuals globally, which confers a substantial burden to the health-care system.2,3 Type 2 diabetes mellitus (T2DM) is a prevalent metabolic disorder characterized by obesity and chronic insulin resistance.4 Globally, the number of people with diabetes mellitus has quadrupled in the past three decades, and diabetes mellitus represents the ninth major cause of death. T2DM comprises about 90% of all diabetic cases.5
Nearly 20 years of research found that T2DM and HF ordinarily occur concomitantly, and each disease independently increases the risk for the other.6,7 On one hand, the prevalence of HF in patients with T2DM (9–22%) is 4 times higher than the general population.6,8 And among patients with HF, those with diabetes have a higher risk of mortality and hospitalization for HF than those without diabetes.9,10 On the other hand, in many HF trials, the incidence of diabetes in patients with heart failure was significantly higher than in the general population during follow-up and observation.6,11,12 Recently numerous studies have attempted to uncover the mechanisms behind the association between heart failure and diabetes. Causing left ventricular concentric remodeling and damaging cardiac microcirculation could be the mechanism of T2DM inducing HF,13–15 and HF could further promote systemic insulin resistance by increasing sympathetic tone and stress-dependent perturbation within metabolic pathways.16 However, the potential mechanisms of the two diseases remain far from being clarified. Simultaneously, some advances in drugs have also suggested there may be a latent common pathway between HF and T2DM.6 For example, hypoglycemic drug SGLT-2 can improve the prognosis of patients with heart failure regardless of whether complicated with diabetes.7,17 Hence, there may be latent common biological mechanisms between HF and T2DM, which are currently unknown, and they will become potential biomarkers and therapeutic directions.
Common transcriptional signatures may provide further insight into the communal molecular mechanisms in HF and T2DM. Therefore, this article aims to reveal the association and potential targets between the two diseases by identifying and analyzing DEGs and hub genes. In this article, we analyzed two gene expression datasets (GSE84796 and GSE95849) which were downloaded from the Gene Expression Omnibus (GEO) to identify the communal DEGs of HF and T2DM. The functional enrichment of DEGs and genes in each four modules were used to uncover the potential molecular mechanisms of HF and T2DM. After that, we constructed a PPI network to screen core modules and hub genes by using STRING database and Cytoscape software. In summary, a total of 236 DEGs, four modules and five hub genes identified in this research contribute to discover the latent biological mechanisms and presents potential biomarkers or therapeutic targets in HF and T2DM.
Materials and Methods
Data Source
GEO (http://www.ncbi.nlm.nih.gov/geo) is a public database containing high throughout sequencing and microarray datasets.18 We screen HF/T2DM datasets based on strict inclusion/exclusion criteria from GEO. The inclusion criteria were as follows: (1) Sporadic heart failure (HF) or type 2 diabetes mellitus (T2DM); (2) Datasets that included patients and healthy controls. Exclusion criteria included: Patients had participated in a clinical trial for drugs or other treatments. Finally, GSE84796 (8 HF patients and 10 controls)19 and GSE95849 (6 T2DM patients and 6 controls)20 were selected from GEO.
Identification of DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) is a web-based tool that based on limma package.18 Differentially expressed genes (DEGs) were identified by GEO2R with the condition that |logFC| > 1 and adj. P-value < 0.05. Subsequently, the overlap DEGs among HF and T2DM datasets were detected and visualized by the online Venn software.
Enrichment Analyses of DEGs
GO and KEGG pathway enrichment analyses were performed to elucidate the biological characteristics of the overlapping DEGs by the online tool DAVID (https://david.ncifcrf.gov/).21 KOBAS 3.0 (http://kobas.cbi.pku.edu.cn) was used for further pathway enrichment analyses from 5 pathway databases (KEGG PATHWAY, PID, BioCyc, Reactome and Panther), and P-value less than 0.05 was considered as statistically significant.22 The genes in four modules were also analyzed in the same way.
Protein–Protein Interaction Network Construction and Module Analysis
Protein–protein interaction (PPI) network was constructed by STRING (http://string-db.org, version 11.0) database with the threshold of the combined score >0.4,23 and the interaction networks were visualized by the Cytoscape (version 3.7.0).24 In addition, MCODE plug-in was used to screen densely interconnected modules in the PPI network with default parameters.25
Hub Genes Selection and Analyses
CytoHubba plug-in in Cytoscape was used to select hub genes in the PPI network.26 We randomly select five of the 12 algorithms in cytoHubba plug-in, and take the intersection of the five algorithms results to determine hub genes. Subsequently, the biological processes enrichment analyses for the genes were conducted by Metascape (https://metascape.org), which is an online platform specialized in comprehensive gene annotation and analysis resource,27 and the enrichment analysis of pathway was through KOBAS 3.0. GeneMANIA (http://www.genemania.org/), a convenient web platform for analyzing gene lists and predicting gene function, was used to analyze and construct a network of hub genes and their co-expression genes.28 Finally, Drug–Gene Interaction database (DGIdb) 3.0 (http://www.dgidb.org/), which used to predict drug-gene pairs, was applied here to predict drugs based on the hub genes.29 After that, we formed the network map of drug–gene interaction by Cytoscape.
Construction of mRNA–miRNA Regulatory Network
Mirwalk, a convenient online database that mainly focuses on miRNA–target interactions,30 was adopted here to predict corresponding miRNA of hub genes by the screening condition that the predicted miRNA could be verified by experiments or other databases. We construct the regulatory network by Cytoscape based on the prediction of mRNA–miRNA.
Results
Identification of DEGs
Two gene expression datasets (GSE84796 and GSE95849) were selected in this research (Figure 1) based on inclusion/exclusion criteria. Subsequently, we used GEO2R to identify the DEGs in the two datasets with |logFC| > 1 and adj. P-value < 0.05. Compared to the controls, 1742 genes (1243 upregulated and 499 downregulated genes) were identified as DEGs in patients with HF (Figure 2A and Supplementary Table 1). Meantime, 2821 DEGs (2697 upregulated and 124 downregulated genes) were screened in patients with T2DM (Figure 2B and Supplementary Table 2). A total of 236 genes overlapped between GSE84796 and GSE95849 datasets as shown in the Venn diagram, consisting of 233 upregulated genes and three downregulated genes (Figure 2C and D).
 |
Figure 1 Flow diagram of the study design. |
Analysis of the Functional Characteristics of DEGs
GO enrichment analysis was used for uncovering the biological roles of the communal DEGs, and results were divided into three functional categories, including biological processes (BP), cell component (CC), and molecular function (MF) (Figure 3A and B). In terms of BP, DEGs were mainly involved in innate immune response (GO:0045087), signal transduction (GO:0007165), immune response (GO:0006955) and inflammatory response (GO:006954). In the CC group, the genes were mainly enriched in plasma membrane (GO:0005886), integral component of plasma membrane (GO:0005887), external side of plasma membrane (GO:0009897) and immunological synapse (GO:0001772). For MF, DEGs were significantly enriched in receptor activity (GO:0004872), receptor binding (GO:0005102), non-membrane spanning protein tyrosine kinase activity (GO:0004715) and G-protein coupled purinergic nucleotide receptor activity (GO:0045028). According to the KEEG pathway analysis results from online database DAVID6.8, DEGs were mainly involved in Chemokine signaling pathway (hsa04062), B cell receptor signaling pathway (hsa04662), Cytokine–cytokine receptor interaction (hsa04060) and Fc gamma R-mediated phagocytosis (hsa04666) (Figure 3C). These results indicate that immune-related play an important role in the communal mechanisms of HF and T2DM.
 |
Figure 3 The function analysis of communal DEGs. (A) The GO enrichment analyses of communal DEGs with P-value and (B) gene count. (C) The pathway analysis of communal DEGs via KOBAS 3.0. |
Protein–Protein Interaction Network Construction and Module Analysis
Based on STRING database, the PPI network of DEGs was constructed with combined scores greater than 0.4 and visualized by Cytoscape software, which contained 178 nodes and 1013 edges (Figure 4 and Supplementary Table 3). Four important modules are obtained from PPI network by MCODE plugin. Then, we use online database KOBAS 3.0 to analyze the pathways enrichment of the genes in each module. As a result, genes in each module all clustered in immune-related areas. In the module A, genes were mainly enriched in Immune System (R-HSA-168256) and Innate Immune System (R-HSA-168249) (Figure 5A). Genes in the module B were mainly enriched in Signal Transduction (R-HSA-162582) and G alpha (i) signalling events (R-HSA-418594) (Figure 5B). In the module C, genes were mainly enriched in Immune System (R-HSA-168256) and Interleukin receptor SHC signaling (R-HSA-912526) (Figure 5C). Genes in the module D were mainly enriched in Immune System (R-HSA-168256) and Innate Immune System (R-HSA-168249) (Figure 5D).
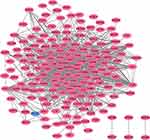 |
Figure 4 Based on database STRING and Cytoscape software, PPI networks of the DEGs were constructed. The red point represents upregulated genes, and blue point represents downregulated genes. |
Hub Gene Selection and Analysis
Five classification methods (BottleNeck, Closeness, MNC, Radiality, Stress) in cytoHubba were used to screen hub genes in this research and the top 10 genes selected by each classification methods are listed Table 1. After that, we determine five central genes by overlapping the top 10 genes in five classification methods (Figure 6).
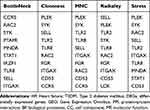 |
Table 1 The Top 10 Hub Genes Rank in cytoHubba |
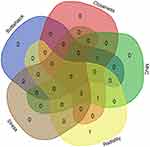 |
Figure 6 Five hub genes were identified by overlapping the first 10 genes in the five classification methods of cytoHubba. |
As expected, functional annotation obtained from Metascape suggested that hub genes were mainly enriched in myeloid leukocyte activation (GO:0002274), leukocyte migration (GO:0050900) and regulation of protein transport (GO:0051223) (Figure 7A). The pathway analyses of the hub genes were conducted using KOBAS 3.0 and pathways with the top four P-values were Innate Immune System (R-HSA-168249), Immune System (R-HSA-168256), Hemostasis (R-HSA-109582) and GPVI-mediated activation cascade (R-HSA-114604) (Figure 7B). Similarly, these results emphasize the important role of Immune System in the HF and T2DM. Besides, a network of the hub genes and their co-expression genes was analyzed by GeneMANIA online platform (Figure 7C). The six genes showed the complex PPI network with the Physical interactions of 67.64%, Co-expression of 13.50%, Co-localization of 6.17%, Predicted of 6.35%, Pathway of 4.35%, Genetic Interactions of 1.40%, and Shared protein domains of 0.59%. Finally, 47 drug–gene interaction pairs were predicted by DGIdb and visualized by Cytoscape, which contained five hub genes (SYK, TLR8, SELL, RAC2 and ITGAX) and 47 drugs (Figure 8). These results may provide clues to therapeutic targets of HF and T2DM.
mRNA–miRNA Regulation Network Construction
Based on the MiRwalk databases predictions of six hub genes, we obtained a total of 109 miRNAs which could be verified by experiments or other databases. After that, a regulatory network of hub genes and predicted miRNAs was constructed by Cytoscape software (Figure 9). These results uncover the potential relationship and mutual regulation between hub genes.
 |
Figure 9 mRNA–miRNA regulation network of hub genes was constructed by MiRwalk. |
Discussion
In this study, we identified 236 overlapping DEGs in both HF and T2DM, of which 233 were upregulated and three were downregulated genes. GO and pathway analysis showed DEGs are mainly enriched in immune response and inflammatory response. Subsequently, we performed pathway analysis on the genes in the four modules respectively, and we found that all four modules were related to immunity. After that, five hub genes (SYK, SELL, RAC2, TLR8 and ITGAX) were found in the PPI network. Functional annotation shows that hub genes are mainly involved in immune-related. Finally, we analysed a network of the hub genes and their co-expression genes, predicted drug–gene interactions and construct mRNA–miRNA Regulation Network.
Interestingly, the functional enrichment of DEGs, genes in each four modules and Hub genes all pointed to immune-related, suggesting that the potential common pathway between T2DM and HF could lie in immune system. Recent 20 years of studies have provided preliminary evidence for the immune mechanism and treatment of T2DM and HF. Studies have presented that immune response activating mechanisms in the heart inducing cardiac adverse remodelling and causing left ventricular dysfunction.31,32 Meanwhile, some experiments have found that elevated levels of circulating pro-inflammatory biomarkers in patients with HF correlate with disease severity and prognosis.33,34 All the above studies suggest that adaptive immunity and innate immunity play an important role in the development of HF. Hence, the public has invested a great deal of effort in the immune modulation therapy of HF in the past 10 years, but the results so far have been disappointing and controversial. This reality might be explained by the complex nature of the interplay between immune cells and their pro- and anti-inflammatory mediators.33,35 Many recent studies have made good progress in the immune mechanism of T2DM. Altered proliferation, function, or infiltration of components of adaptive immunity and innate immunity is critical in the progression of T2DM, such as increased number of CD45+ T cell, leukocyte shift toward a pro-inflammatory phenotype, and a reduction in the number of suppressive regulatory T cells and protective NK cells.4,36 Recently several studies focused on the immune effects of hypoglycemic drugs or the use of immune system modulators to improve glucose and lipid metabolism and the results were proved to be meaningful.36,37 In summary, immune-related directions may be the potential common biomarkers and therapeutic directions of HF and T2DM, and more experiments are needed to uncover the immune-related associations of HF and T2DM.
SYK (spleen-associated tyrosine kinase), this gene encodes a member of the family of non-receptor type Tyr protein kinases. This protein is widely expressed in hematopoietic cells and is involved in coupling activated immunoreceptors to downstream signalling events that mediate diverse cellular responses, including proliferation, differentiation and phagocytosis.38–40 The relationship between SYK and diabetes development was found to be mainly related to immune receptors. Konigsberger et al found that disrupting SYK expression resulted in the alteration in BCR signalling quality which could cause the generation of anti-insulin antibodies.41 In addition, some experiments revealed that cardiovascular events in diabetic patients were closely associated with SYK, mainly related to platelet dysfunction caused by SYK. Yamagishi et al reported that hyperglycaemia increasing phosphorylation of SYK, which in turn result in platelet dysfunction, played an important role in the pathogenesis of diabetic micro- and microangiopathies.42 Meanwhile, Calverley et al found that increased platelet FCR-mediated sensitivity to collagen, which could be induced by the enhancing levels of tyrosine-phosphorylated SYK in diabetic platelets, may result in cardiovascular morbidity and mortality among patients with diabetes.43 As above, there is no denying that SYK is a potential target for heart failure and diabetes.
TLR8 (Toll-like receptor 8) is a member of the Toll-like receptor (TLR) family. As we all known, Toll-like receptors (TLRs) play essential roles in generating innate immune responses, and then recognized by ligand activating signalling cascades. Beyond this, recent evidence suggests that it plays even broader roles in pathogen-specific adaptive immune responses and immunity.44 TLR8 profiled over expression in enterovirus-associated dilated cardiomyopathy and chronic chagasic cardiomyopathy, which may eventually develop into HF.45,46 Meanwhile, TLR8 is elevated expression in adipose tissue among patients with T2DM.47 Thus, TLR8 could be an immune marker of metabolic inflammation in HF and T2DM. Similar to TLR8, ITGAX (integrin subunit alpha X) is a macrophage/dendritic cell marker, also could be used as an indicator. Some experiments have found that islet inflammation could promote beta cell dysfunction in T2DM via islet inflammation with increased expression of ITGAX.48,49 Unfortunately, the association between ITGAX and HF has not yet been found. To sum up, TLR8 and ITGAX could be potential biomarker for HF and T2DM, but more experiments and research are needed before it.
SELL (selectin L) encodes a cell surface adhesion molecule that belongs to a family of adhesion/homing receptors. The gene product is required for binding and subsequent rolling of leucocytes on endothelial cells, facilitating their migration into secondary lymphoid organs and inflammation sites.50 Some experiments have shown that SELL combining with its monoclone antibody and ligand can cause cellular morphologic change, cellular framework rearrangement, oxygen respiration explosion, increasing IL-8 and TNF-α gene expression. Based on the fact that TNF-α is one of the main factors that induces insulin resistance, we speculate SELL may take part in the development of the insulin resistance.51,52 Moreover, in some cases, measuring serum adhesion molecules may help predict, early diagnose, prevent and monitor the treatment complications of type 2 diabetes.52–54 Currently, there is no evidence of association between SELL and HF, and most of the evidence focuses on coronary heart disease. The high expression of SELL can damage cells and platelets, and then induce coronary atherosclerosis or even acute coronary syndrome.55 However, the role of SELL in ischemic heart failure still needs to be further clarified. RCA2 encodes a member of the Ras superfamily of small guanosine triphosphate (GTP)-metabolizing proteins.56 RAC2 was found to be involved in the biological process of insulin-secreting INS-1 cells against cytokine-induced apoptosis.57 However, there have been few studies and no clear evidence of an association between RAC2 and HF or T2DM. More experiments are needed to clarify the relationship between RCA2 and HF or T2DM.
The limitations of our study are as follows: the sample size of our study was relatively small and external validation is needed to confirm our results. Hence, our conclusions need to be validated by large sample sizes and further mechanism experiments.
Conclusion
In conclusion, the communal DEGs and modules identified in our study and subsequent analyses reveal potential common mechanisms of HF and T2DM. Besides, five hub genes (SYK, SELL, RAC2, TLR8, ITGAX) have been identified, which can be used as biomarkers or therapeutic targets for HF and T2DM.
Ethics Statement
GEO belongs to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data, so there are no ethical issues and other conflicts of interest. In all, our study is exempt from ethical approval.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Clinical trial ability improvement project of cardiovascular professional group in the First Affiliated Hospital of Soochow University (No:201900180019).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: the task force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J. 2016;37(27):2129–2200.
2. Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol. 2016;13(6):368–378. doi:10.1038/nrcardio.2016.25
3. Ponikowski P, Anker SD, AlHabib KF, et al. Heart failure: preventing disease and death worldwide. ESC Heart Fail. 2014;1(1):4–25. doi:10.1002/ehf2.12005
4. Berbudi A, Rahmadika N, Tjahjadi AI, Ruslami R. Type 2 diabetes and its impact on the immune system. Curr Diabetes Rev. 2020;16(5):442–449. doi:10.2174/1573399815666191024085838
5. Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14(2):88–98. doi:10.1038/nrendo.2017.151
6. Dunlay SM, Givertz MM, Aguilar D, et al. Type 2 diabetes mellitus and heart failure: a scientific statement from the American Heart Association and the Heart Failure Society of America: this statement does not represent an update of the 2017 ACC/AHA/HFSA heart failure guideline update. Circulation. 2019;140(7):e294–e324. doi:10.1161/CIR.0000000000000691
7. Seferović PM, Fragasso G, Petrie M, et al. Sodium-glucose co-transporter 2 inhibitors in heart failure: beyond glycaemic control. A position paper of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail. 2020;22(9):1495–1503. doi:10.1002/ejhf.1954
8. Nichols GA, Gullion CM, Koro CE, Ephross SA, Brown JB. The incidence of congestive heart failure in type 2 diabetes: an update. Diabetes Care. 2004;27(8):1879–1884. doi:10.2337/diacare.27.8.1879
9. Aguilar D, Solomon SD, Køber L, et al. Newly diagnosed and previously known diabetes mellitus and 1-year outcomes of acute myocardial infarction: the VALsartan in Acute myocardial iNfarcTion (VALIANT) trial. Circulation. 2004;110(12):1572–1578. doi:10.1161/01.CIR.0000142047.28024.F2
10. MacDonald MR, Petrie MC, Varyani F, et al. Impact of diabetes on outcomes in patients with low and preserved ejection fraction heart failure: an analysis of the Candesartan in Heart failure: Assessment of Reduction in Mortality and morbidity (CHARM) programme. Eur Heart J. 2008;29(11):1377–1385. doi:10.1093/eurheartj/ehn153
11. Preiss D, Zetterstrand S, McMurray JJ, et al. Predictors of development of diabetes in patients with chronic heart failure in the Candesartan in Heart Failure Assessment of Reduction in Mortality and Morbidity (CHARM) program. Diabetes Care. 2009;32(5):915–920. doi:10.2337/dc08-1709
12. Preiss D, van Veldhuisen DJ, Sattar N, et al. Eplerenone and new-onset diabetes in patients with mild heart failure: results from the Eplerenone in Mild Patients Hospitalization and Survival Study in Heart Failure (EMPHASIS-HF). Eur J Heart Fail. 2012;14(8):909–915. doi:10.1093/eurjhf/hfs067
13. Levelt E, Rodgers CT, Clarke WT, et al. Cardiac energetics, oxygenation, and perfusion during increased workload in patients with type 2 diabetes mellitus. Eur Heart J. 2016;37(46):3461–3469. doi:10.1093/eurheartj/ehv442
14. Levelt E, Pavlides M, Banerjee R, et al. Ectopic and visceral fat deposition in lean and obese patients with type 2 diabetes. J Am Coll Cardiol. 2016;68(1):53–63. doi:10.1016/j.jacc.2016.03.597
15. Lehrke M, Marx N. Diabetes mellitus and heart failure. Am J Cardiol. 2017;120(1s):S37–s47. doi:10.1016/j.amjcard.2017.05.014
16. Doehner W, Frenneaux M, Anker SD. Metabolic impairment in heart failure: the myocardial and systemic perspective. J Am Coll Cardiol. 2014;64(13):1388–1400. doi:10.1016/j.jacc.2014.04.083
17. Hallow KM, Helmlinger G, Greasley PJ, McMurray JJV, Boulton DW. Why do SGLT2 inhibitors reduce heart failure hospitalization? A differential volume regulation hypothesis. Diabetes Obes Metab. 2018;20(3):479–487. doi:10.1111/dom.13126
18. Barrett T, Wilhite S, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(D1):D991–995. doi:10.1093/nar/gks1193
19. Laugier L, Frade A, Ferreira F, et al. Whole-genome cardiac DNA methylation fingerprint and gene expression analysis provide new insights in the pathogenesis of chronic Chagas disease cardiomyopathy. Clin Infect Dis. 2017;65(7):1103–1111. doi:10.1093/cid/cix506
20. Luo L, Zhou WH, Cai JJ, et al. Gene expression profiling identifies downregulation of the neurotrophin-MAPK signaling pathway in female diabetic peripheral neuropathy patients. J Diabetes Res. 2017;2017:8103904. doi:10.1155/2017/8103904
21. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
22. Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(1):W720–724. doi:10.1093/nar/gkl167
23. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(D1):D808–815. doi:10.1093/nar/gks1094
24. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi:10.1093/bioinformatics/btq675
25. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4(1):2. doi:10.1186/1471-2105-4-2
26. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl S4):S11. doi:10.1186/1752-0509-8-S4-S11
27. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
28. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl_2):W214–220. doi:10.1093/nar/gkq537
29. Cotto KC, Wagner AH, Feng YY, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–d1073. doi:10.1093/nar/gkx1143
30. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. doi:10.1371/journal.pone.0206239
31. Mortensen RM. Immune cell modulation of cardiac remodeling. Circulation. 2012;125(13):1597–1600. doi:10.1161/CIRCULATIONAHA.112.097832
32. Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science. 2013;339(6116):161–166. doi:10.1126/science.1230719
33. Zhang Y, Bauersachs J, Langer HF. Immune mechanisms in heart failure. Eur J Heart Fail. 2017;19(11):1379–1389. doi:10.1002/ejhf.942
34. Anand IS, Latini R, Florea VG, et al. C-reactive protein in heart failure: prognostic value and the effect of valsartan. Circulation. 2005;112(10):1428–1434. doi:10.1161/CIRCULATIONAHA.104.508465
35. Mielniczuk LM, Baughman KL. Immune modulation therapy in heart failure. Congest Heart Fail. 2006;12(2):
36. Zhou T, Hu Z, Yang S, Sun L, Yu Z, Wang G. Role of adaptive and innate immunity in type 2 diabetes mellitus. J Diabetes Res. 2018;2018:7457269. doi:10.1155/2018/7457269
37. Xu X, Lin S, Chen Y, et al. The effect of metformin on the expression of GPR109A, NF-κB and IL-1β in peripheral blood leukocytes from patients with type 2 diabetes mellitus. Ann Clin Lab Sci. 2017;47(5):556–562.
38. Yin G, Yang X, Li Q, Guo Z. SYK protects cardiocytes against anoxia and hypoglycemia-induced injury in ischemic heart failure. Mol Immunol. 2017;91:35–41. doi:10.1016/j.molimm.2017.08.018
39. Wei W, Li XX, Xu M. Inhibition of vascular neointima hyperplasia by FGF21 associated with FGFR1/Syk/NLRP3 inflammasome pathway in diabetic mice. Atherosclerosis. 2019;289:132–142. doi:10.1016/j.atherosclerosis.2019.08.017
40. Liu L, Pang X, Shang W, Feng G, Wang Z, Wang J. miR-136 improves renal fibrosis in diabetic rats by targeting down-regulation of tyrosine kinase SYK and inhibition of TGF-β1/Smad3 signaling pathway. Ren Fail. 2020;42(1):513–522. doi:10.1080/0886022X.2020.1764854
41. Königsberger S, Prodöhl J, Stegner D, et al. Altered BCR signalling quality predisposes to autoimmune disease and a pre-diabetic state. EMBO J. 2012;31(15):3363–3374. doi:10.1038/emboj.2012.169
42. Yamagishi SI, Edelstein D, Du XL, Brownlee M. Hyperglycemia potentiates collagen-induced platelet activation through mitochondrial superoxide overproduction. Diabetes. 2001;50(6):1491–1494. doi:10.2337/diabetes.50.6.1491
43. Calverley DC, Hacker MR, Loda KA, et al. Increased platelet Fc receptor expression as a potential contributing cause of platelet hypersensitivity to collagen in diabetes mellitus. Br J Haematol. 2003;121(1):139–142. doi:10.1046/j.1365-2141.2003.04233.x
44. Lanzavecchia A, Sallusto F. Toll-like receptors and innate immunity in B-cell activation and antibody responses. Curr Opin Immunol. 2007;19(3):268–274. doi:10.1016/j.coi.2007.04.002
45. Satoh M, Akatsu T, Ishikawa Y, Minami Y, Takahashi Y, Nakamura M. Association between toll-like receptor 8 expression and adverse clinical outcomes in patients with enterovirus-associated dilated cardiomyopathy. Am Heart J. 2007;154(3):581–588. doi:10.1016/j.ahj.2007.05.010
46. Pereira NS, Queiroga TBD, Nunes DF, et al. Innate immune receptors over expression correlate with chronic chagasic cardiomyopathy and digestive damage in patients. PLoS Negl Trop Dis. 2018;12(7):e0006589. doi:10.1371/journal.pntd.0006589
47. Ahmad R, Kochumon S, Thomas R, Atizado V, Sindhu S. Increased adipose tissue expression of TLR8 in obese individuals with or without type-2 diabetes: significance in metabolic inflammation. J Inflamm. 2016;13(1):38. doi:10.1186/s12950-016-0147-y
48. Meier DT, Morcos M, Samarasekera T, Zraika S, Hull RL, Kahn SE. Islet amyloid formation is an important determinant for inducing islet inflammation in high-fat-fed human IAPP transgenic mice. Diabetologia. 2014;57(9):1884–1888. doi:10.1007/s00125-014-3304-y
49. Hannibal TD, Schmidt-Christensen A, Nilsson J, Fransén-Pettersson N, Hansen L, Holmberg D. Deficiency in plasmacytoid dendritic cells and type I interferon signalling prevents diet-induced obesity and insulin resistance in mice. Diabetologia. 2017;60(10):2033–2041. doi:10.1007/s00125-017-4341-0
50. Ivetic A, Hoskins Green HL, Hart SJ. L-selectin: a major regulator of leukocyte adhesion, migration and signaling. Front Immunol. 2019;10:1068.
51. Chen C, Ba X, Xu T, Cui L, Hao S, Zeng X. c-Abl is involved in the F-actin assembly triggered by L-selectin crosslinking. J Biochem. 2006;140(2):229–235. doi:10.1093/jb/mvj149
52. Liu J, Liu JX, Xu SN, et al. Association of P213S polymorphism of the L-selectin gene with type 2 diabetes and insulin resistance in Chinese population. Gene. 2012;509(2):286–290. doi:10.1016/j.gene.2012.07.086
53. Kamiuchi K, Hasegawa G, Obayashi H, et al. Leukocyte-endothelial cell adhesion molecule 1 (LECAM-1) polymorphism is associated with diabetic nephropathy in type 2 diabetes mellitus. J Diabetes Complicat. 2002;16(5):333–337. doi:10.1016/S1056-8727(01)00226-4
54. Sokup A. [Adhesion molecules in type 2 diabetes]. Przegl Lek. 2005;62(4):234–237. Polish
55. Sandoval-Pinto E, Padilla-Gutiérrez JR, Hernández-Bello J, et al. Influence of haplotypes, gene expression and soluble levels of L-selectin on the risk of acute coronary syndrome. Gene. 2017;625:31–41. doi:10.1016/j.gene.2017.05.005
56. Arrington ME, Temple B, Schaefer A, Campbell SL. The molecular basis for immune dysregulation by the hyperactivated E62K mutant of the GTPase RAC2. J Biol Chem. 2020;295(34):12130–12142. doi:10.1074/jbc.RA120.012915
57. Fløyel T, Mirza AH, Kaur S, et al. The Rac2 GTPase contributes to cathepsin H-mediated protection against cytokine-induced apoptosis in insulin-secreting cells. Mol Cell Endocrinol. 2020;518:110993. doi:10.1016/j.mce.2020.110993
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.