Back to Journals » Journal of Inflammation Research » Volume 16
An in-Depth Exploration of the Genetic Interaction Network Between Ferroptosis and Acute Pancreatitis
Authors Li J, Jia Y, Cao F , Wang G, Li F
Received 20 July 2023
Accepted for publication 3 October 2023
Published 6 October 2023 Volume 2023:16 Pages 4425—4439
DOI https://doi.org/10.2147/JIR.S431601
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Jie Li,1,2 Yuchen Jia,1,2 Feng Cao,1,2 Gang Wang,3 Fei Li1,2
1Department of General Surgery, Xuanwu Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Clinical Center for Acute Pancreatitis, Capital Medical University, Beijing, People’s Republic of China; 3Department of Pancreatic and Biliary Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, People’s Republic of China
Correspondence: Fei Li; Gang Wang, Email [email protected]; [email protected]
Background: Ferroptosis plays an important role in a variety of disease processes and is equally important in pancreatic diseases. However, the role of ferroptosis-related genes (FRGs) in acute pancreatitis (AP) remains unknown, and their specific potential mechanisms still need to be explored extensively.
Methods: AP-related gene microarray data were obtained from the GEO database, while FRGs were obtained from the ferroptosis database (FerrDb). Differentially expressed genes (DEGs) were screened by the “limma” package, and GSEA was performed. The corresponding ferroptosis-related differentially expressed genes (FRDEGs) were screened, and GO and KEGG pathway analyses were performed. A PPI network was constructed to identify hub FRDEGs by CytoHubba, MCODE and CTD scores. Transcription factors and miRNAs predicted using the NetworkAnalyst database were used to establish the regulatory network. Immune cell infiltration analysis was performed by the R package “ssGSEA” algorithm. The hub genes were validated by transcriptome sequencing of AP model mice and immunohistochemistry in rats and mice.
Results: A total of 82 FRDEGs were screened, and these genes were mainly associated with ferroptosis, hypoxic response, autophagy, mitophagy and immune inflammation. However, we also found that these genes are also jointly involved in other cell death modalities, such as apoptosis and necroptosis. Further analysis obtained 7 hub genes from 82 genes, and single-sample gene set enrichment analysis (ssGSEA) showed that the hub genes are closely associated with the infiltration of specific immune cells and the activation of immune pathways.
Conclusion: This study reveals the complex functions and important roles of ferroptosis-related genes in AP and provides gene targets for further studies of AP.
Keywords: acute pancreatitis, ferroptosis, mitophagy, autophagy, inflammation
Introduction
AP is a common surgical acute abdominal condition that is often complicated by local complications. Although significant progress has been made on elucidating the pathogenesis of AP, the specific mechanism is still not very clear.1,2 Pancreatic acinar cell death is considered to be one of the important factors in the development of AP and a key factor in the disease progression of AP. Therefore, the inhibition of acinar cell death is the focus of the prevention and treatment of AP.
In recent years, ferroptosis, a new form of cell death, has aroused heated discussion.3–7 Ferroptosis is markedly different from necrosis, apoptosis and autophagy in terms of cell morphology and function.4,8 Ferroptosis emphasizes the importance of iron ions to ROS. After Fe2+ aggregates, it can oxidize lipids and produce large amounts of ROS, causing cells to die. There is also evidence that there is a process of accumulation of iron ions, a decrease in GSH and the accumulation of lipid peroxides in AP.9,10 Therefore, whether there is such a mode of death in AP and whether a therapeutic effect on AP can be achieved by regulating ferroptosis is a topic worthy of further study.
Currently, a number of researchers have gradually linked ferroptosis and AP to explore the prevention of inflammatory injury in the pancreas after inhibiting ferroptosis.11–15 Moreover, it is not only limited to pancreatic injury in AP but is also involved in the kidney injury and lung injury caused by AP.16–18 These findings emphasize the important role of ferroptosis in the AP process. Ultimately, it was found that ferroptosis and AP are closely associated and jointly affect the AP disease process in several ways.
Materials and Methods
Animals and Reagents
C57BL/6 mice (SPF, 6 weeks old, 20 ± 2 g) and Wistar rats (SPF, 200–250 g) were purchased from the WeiTongLiHua Experimental Animal Technical Company (Beijing, China). Mice and rats were provided standard feed and water in an environmentally controlled room (18–21 °C, 40–60% relative humidity, 12 h light/dark cycle). After acclimatization for one week, both mice and rats were fasted overnight before the experiments. The animal experiments were approved by the Ethics Committee of Xuanwu Hospital of Capital Medical University and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Caerulein was purchased from MCE (Shanghai, China). Sodium taurocholate (Na-TC) was purchased from Sigma‒Aldrich (St. Louis, MO, USA). Antibodies (anti-Stat3, anti-Jun, anti-Il6, anti-Rela, anti-Hmox1, anti-Hif1a and anti-Sqstm1) were purchased from Boster (Wuhan, China).
Animal Models
Mice were randomly divided into 2 groups: the Control and AP groups. Each mouse was weighed individually, and caerulein was injected intraperitoneally at a dose of 50 μg/kg every 1 hour 8 consecutive times. The Control group was injected with saline. One hour after the last injection, the mice were euthanized, and pancreatic tissue was collected.
Rats were anesthetized by intraperitoneal injection of sodium pentobarbital (40 mg/kg). A midline open laparotomy was then performed to ligate the distal pancreaticobiliary duct. AP was induced by retrograde injection of 3.5% Na-TC (0.15 mL/100 g) into the pancreaticobiliary duct. Sham rats underwent open abdominal surgery only.
RNA Sequencing
Total RNA was extracted from the fresh mouse pancreas using TRIzol Reagent according the manufacturer’s instructions (Magen). RNA samples were detected based on the A260/A280 absorbance ratio with a Nanodrop ND-2000 system (Thermo Scientific, USA), and the RIN of RNA was determined by an Agilent Bioanalyzer 4150 system (Agilent Technologies, CA, USA). Only qualified samples will be used for library construction. Paired-end libraries were prepared using a ABclonal mRNA-seq Lib Prep Kit (ABclonal, China) following the manufacturer’s instructions. The mRNA was purified from 1 μg total RNA using oligo (dT) magnetic beads followed by fragmentation carried out using divalent cations at elevated temperatures in ABclonal First Strand Synthesis Reaction Buffer. Subsequently, first-strand cDNAs were synthesized with random hexamer primers and Reverse Transcriptase (RNase H) using mRNA fragments as templates, followed by second-strand cDNA synthesis using DNA polymerase I, RNAseH, buffer, and dNTPs. The synthesized double stranded cDNA fragments were then adapter ligated for preparation of the paired-end library. Adaptor-ligated cDNA were used for PCR amplification. PCR products were purified (AMPure XP system) and library quality was assessed on an Agilent Bioanalyzer 4150 system. Finally, the library preparations were sequenced on an Illumina Novaseq 6000 and 150 bp paired-end reads were generated.
Data Access
Three datasets (GSE3644, GSE65146, and GSE109227) were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), and all three datasets contained mouse normal pancreas samples and AP pancreas samples. After our final selection, GSE3644 contained 3 normal samples and 3 AP samples, GSE65146 contained 5 normal samples and 9 AP samples, and GSE109227 contained 5 normal samples and 6 AP samples. In addition, 212 ferroptosis genes were obtained from the FerrDb database (http://www.zhounan.org/ferrdb/).
Differential Expression Analysis
DEGs were analyzed and filtered from the normal and AP samples in three datasets using the R “limma” package. The filtering criteria were adjusted P value ≤ 0.05 and absolute value of the logarithmic fold change |log2FC| ≥ 1. Next, we intersected the filtered differential genes with the set of ferroptosis genes to obtain FRDEGs.
Functional Enrichment and Signaling Pathways
Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and single-gene gene set enrichment analysis (GSEA) were performed using R (clusterProfiler, enrichplot).
Construction of PPI and TF-miRNA Networks
PPI networks were analyzed and constructed using the STRING database (https://cn.string-db.org/), and transcription factors (TFs) and miRNAs of selected genes were predicted using NetworkAnalyst (https://www.networkanalyst.ca/). The networks were filtered and visualized using Cytoscape software.
Correlation Analysis of Hub Genes with Disease and Immunity
The CTD database was used to predict the relationship between genes and acute pancreatitis. The R package “ssgsea” algorithm was used to analyze the merged dataset for 28 immune cells and was visualized with heatmaps and boxplots. Correlation analysis was performed between genes and between genes and immune cells.
Histological Staining
Pancreatic tissues of mice and rats were fixed in 4% paraformaldehyde, paraffin-embedded in sections and adhered to slides. Hematoxylin and eosin (H&E) staining was used to examine the extent of inflammation and tissue damage in pancreatic tissues. Two experienced pathologists blindly scored the experimental tissues. The degree of edema, inflammation, hemorrhage, and necrosis of the pancreatic tissue was scored on a scale of 0 to 4 in each of 20 random high magnification views according to the histological scoring criteria reported by Kusske et al19 Paraffin-embedded tissue sections were processed through a series of procedures and stained with antibodies (anti-Stat3, anti-Jun, anti-Il6, anti-Rela, anti-Hmox1, anti-Hif1a and anti-Sqstm1). Images were taken with an Olympus (Tokyo, Japan) camera.
Statistical Analysis
Comparisons between the two groups were made using Student’s t-test. Differential gene analysis was performed using the “limma” package of R software. Pearson’s correlation analysis was used to reveal the relationships among the 7 hub genes. Cytoscape was used to visualize the PPI and TF-miRNA networks. p < 0.05 was considered significant. All analyses were performed in R.
Results
DEGs in AP and Functional Enrichment Analysis
In this study, we selected three AP-related datasets (GSE3644, GSE65146, and GSE109227) from the GEO database for analysis. We performed differential gene analysis on the three datasets, and the results showed that there were 383 differential genes in the GSE3644 dataset, including 302 upregulated genes and 81 downregulated genes in the AP group compared with the control group (Figure 1A and D). There were 4797 differential genes in the GSE65146 dataset, including 3397 upregulated genes and 1400 downregulated genes (Figure 1B and E). There were 1541 differential genes in the GSE109227 dataset, including 1249 upregulated genes and 292 downregulated genes (Figure 1C and F). The DEGs were visualized as volcano plots and heatmaps.
Next, to explore the association between AP and ferroptosis, we performed GSEA of differentially expressed genes (Figure 1G–I). The GSEA results showed corresponding enrichment in the ferroptosis pathway and its related metabolic pathways, including arachidonic acid metabolism, fatty acid degradation, fatty acid metabolism, glutathione metabolism, oxidative phosphorylation, and phosphatidylinositol signaling system.
Determination of Ferroptosis-Related DEGs
Ferroptosis-related genes were downloaded from the FerrDb database and intersected with the DEGs of the three datasets. The overlapping genes were selected as FRDEGs. The results showed that there were 17 FRDEGs in GSE3644, 66 FRDEGs in GSE65146, and 43 FRDEGs in GSE109227. We combined the FRDEGs from each dataset and finally obtained 82 FRDEGs, which were used in the subsequent analysis (Figure 2).
We performed GO enrichment and KEGG pathway analysis on FRDEGs from each dataset and the 82 FRDEGs obtained from the final screening (Figures 2 and 3). The final GO enrichment results showed that terms mainly included oxidative stress, transcriptional regulation, autophagosome formation and ubiquitination of proteins (Figure 3A). KEGG pathways in FRDEGs were mainly ferroptosis pathways, mitophagy, autophagy regulation, immune-related pathways, and some other cell death modalities, such as apoptosis and necroptosis (Figure 3B and C). This evidence suggests that ferroptosis is not only significantly associated with the autophagic process in AP but also a coregulated gene network with other cell death modalities. In addition, ferroptosis is also involved in certain immune regulatory processes.
 |
Figure 3 Enrichment analysis of 82 FRDEGs. (A) GO enrichment analysis of 82 FRDEGs. (B and C) KEGG enrichment analysis of 82 FRDEGs. |
PPI Network Analysis and Hub FRDEG Identification
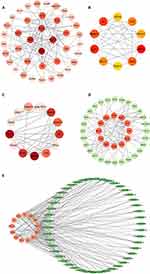
Analysis and construction of PPI networks using the STRING database. Eighty-two FRDEGs were included in the database for analysis, and the network was visualized using Cytoscape software (Figure 4A). Using CytoHubba and the MCODE function of Cytoscape for hub gene screening, we finally screened 10 and 13 hub genes, respectively (Figure 4B and C). Combining the results, we finally obtained 13 hub FRDEGs, including Stat3, Jun, Il6, Mapk14, Rela, Tlr4, Becn1, Hmox1, Hif1a, Sqstm1, Map1lc3a, Wipi1 and Wipi2.
Prediction of Transcription Factors and miRNAs of Hub FRDEGs
NetworkAnalyst was used to predict the transcription factors and miRNAs of the selected 13 genes, and Cytoscape was used to visualize the network. Transcription factor and gene target data were derived from the ENCODE ChIP-seq data. We obtained a network of 40 nodes, 72 edges, and 12 seeds, from which we identified 28 TFs (Figure 4D). Comprehensive experimentally validated miRNA-gene interaction data were collected from TarBase. We obtained a total of 166 nodes, 342 edges, and 13 seeds, from which we obtained 153 miRNAs that were finally visualized by Cytoscape analysis with degree ≥3 (Figure 4E). The final results still need to be verified by experiments.
Correlation of Hub Genes with Disease and Immunity
The CTD database was used to predict the association between hub genes and acute pancreatitis. We obtained scores for the correlation between AP and hub genes from the CTD database. Genes with a score higher than 50% were considered to have a higher association. After screening, we obtained 7 genes, namely, Stat3, Jun, Il6, Rela, Hmox1, Hif1a and Sqstm1 (Figure 5A and B) (Table 1). We analyzed the correlation of these seven genes and found that they all showed a positive correlation. Among them, the correlations between Hif1a and Jun, Il6, and Stat3 (the scores were 0.96, 0.96, and 0.98, respectively), the correlation between Rela and Hmox1 (score of 0.96) and the correlation between Jun and Stat3 (score of 0.98) were high (Figure 5C). Next, we performed an analysis of the respective pathways of these 7 genes to determine whether these genes are correlated with immunity. Finally, we found that these genes are associated with immunity-related pathways, such as the formation of the neutrophil extracellular trap network and the B-cell receptor signaling pathway (Figure 5D–J).
 |
Table 1 Current Research Progress on 7 Hub Genes |
Immune Infiltration Analysis
We analyzed the merged dataset for 28 immune cells using the R package “ssgsea” algorithm and visualized it with heatmaps and boxplots. Finally, the correlations of 7 key genes with 28 immune cells were described in detail. We found that the levels of macrophages, B cells and T cells were elevated to some extent in AP, but there were no significant changes in the levels of neutrophils, which may be caused by the early stage of AP in the pancreatic samples (Figure 6A and B). Correlation analysis revealed that Rela and Hmox1 had the highest correlation in T cells, and Hif1a and Sqstm1 had the highest correlation in B cells. These 7 genes were not significantly correlated in macrophages and neutrophils (Figure 6C).
Validation of Hub Genes
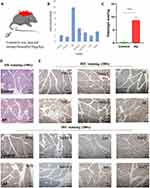
As previously described, mice were administered an intraperitoneal injection of caerulein (8 consecutive injections at 1 hr intervals) to establish a mouse AP model. HE staining was used to evaluate the degree of inflammation in AP (Figure 7A, C and D). The pancreas of mice was transcriptome sequenced and used to validate the expression levels of 7 genes. Finally, we found that the RNA expression levels of all 7 genes were significantly higher in AP mice than in control mice (Log2Foldchange > 1) (Figure 7B). Immunohistochemistry was also used for further validation, and the results were consistent with the previous results (Figure 7E).
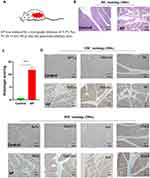
We also constructed a rat model of AP by retrograde injection of 3.5% Na-TC (0.15 mL/100 g) into the pancreaticobiliary duct (Figure 8A). HE staining was used to evaluate the degree of inflammation in AP, and immunohistochemistry was used to verify the expression of 7 genes (Figure 8B–D). The results of the two AP models were consistent.
Discussion
The pathogenesis of AP is complex, and AP is a disease presenting with digestive effects, edema, hemorrhage, and even necrosis of pancreatic tissue caused by a variety of reasons. When AP occurs, the oxidative and antioxidant systems in the body are out of balance, leading to the production and accumulation of large amounts of intracellular ROS. This, in turn, leads to the production of lipid peroxides, resulting in the death of acinar cells.13,20,21 In the pathogenesis of AP, there are multiple modes of acinar cell death, including necrosis, apoptosis, autophagy, and other cell death modes. All of these modes of death affect the progression of AP to different degrees.22 Ferroptosis, however, is different from all these modes. It does not have the morphological features of typical necrosis, such as the swelling of cytoplasm and organelles and rupture of cell membranes, nor does it have the features of traditional apoptosis, such as cell shrinkage, chromatin condensation, formation of apoptotic bodies and disintegration of the cytoskeleton. Unlike autophagy, ferroptosis does not form the classic closed double-layer membrane structure.4,8 These results indicate the specificity of ferroptosis. Currently, the study of ferroptosis in AP is still at a very superficial stage.
Our study delved into the link between AP and ferroptosis and analyzed the pathways associated with FRDEGs in AP. Interestingly, we found that these genes of ferroptosis are correlated with other modes of cell death, including necrosis and apoptosis. This suggests a possible network of ferroptosis regulation in AP that interacts with other cell death modalities. Multiple mechanisms coexist and coregulate each other,23 which ultimately affects the development of AP. In addition, FRDEGs are highly associated with the formation of autophagosomes. We found that FRDEGs are highly correlated not only with macroautophagy but also with mitophagy (an autophagy pathway targeting mitochondria). In recent years, the relationship between autophagy and ferroptosis has also been gaining attention.24–27 Mitophagy, a type of selective autophagy, plays a pivotal role in maintaining mitochondrial homeostasis in response to the removal of damaged mitochondria.28–30 Interestingly, mitophagy is both friend and foe to ferroptosis. In response to mild stress or in the early stages of iron overload, mitophagy may sequester iron in the mitophagosome and reduce the source of ROS for ferroptosis.31 However, extensive mitophagy may provide an additional source of iron to amplify lipid peroxidation and ferroptosis. Thus, our study only illustrates the strong correlation between ferroptosis and mitophagy in AP, and specific details remain to be continuously explored. In this study, we ultimately identified seven genes (Stat3, Jun, Il6, Rela, Hmox1, Hif1a, and Sqstm1) with important regulatory roles in AP, and they are associated with high scoring coefficients for disease. Among these 7 genes, there are some genes that are involved in classic inflammatory pathways, such as the Rela (NF-κB) pathway, Il6/Jak2/Stat3 signaling pathway, and Nrf2/Hmox1 signaling pathway, according to previous studies. The inhibition of these pathways can effectively improve the progression of AP.32–34 Our latest findings may also provide new research directions for a deeper understanding of the mechanisms underlying AP development. Sqstm1 is an important selective autophagy junction protein that plays an important role in regulating autophagy in AP.34 In recent years, Hif1a has been found to regulate autophagy and apoptosis in AP.20,35 In addition, in recent years, Jun/GPX4 has been found to regulate ferroptosis in acinar cells in AP.11 Thus, we are more certain about the multiple functions of these genes, and the discovery of their tight connection with ferroptosis will be of great help in the study of AP. In addition, these genes also have a role in immunity. Among the innate immune cells we focused on in the AP stage, Rela and Hmox1 demonstrated the highest correlation in T cells, and Hif1a and Sqstm1 demonstrated the highest correlation in B cells. These seven genes were not significantly relevant for macrophages and neutrophils.
Conclusion
Our study further revealed a close link between ferroptosis and AP. In this study, we identified seven FRDEGs (Stat3, Jun, Il6, Rela, Hmox1, Hif1a, and Sqstm1) that are closely associated with AP. Furthermore, correlations between ferroptosis and apoptosis, necrosis and autophagy in AP were revealed. Finally, we also found that FRDEGs have a regulatory effect on immune cells. Our study provides an important research target for the study of ferroptosis in AP, as well as a basis for the appropriate treatment of AP.
Acknowledgments
This paper was supported by grants from the Beijing Municipal Science and Technology Commission (No. Z171100001017077, Fei Li).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Gukovsky I, Pandol SJ, Gukovskaya AS. Organellar dysfunction in the pathogenesis of pancreatitis. Antioxid Redox Signal. 2011;15(10):2699–2710. doi:10.1089/ars.2011.4068
2. Everhart JE, Ruhl CE. Burden of digestive diseases in the United States part III: liver, biliary tract, and pancreas. Gastroenterology. 2009;136(4):1134–1144. doi:10.1053/j.gastro.2009.02.038
3. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
4. Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88. doi:10.1038/s41419-020-2298-2
5. Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi:10.1038/ncb3064
6. Doll S, Freitas FP, Shah R, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi:10.1038/s41586-019-1707-0
7. Mao C, Liu X, Zhang Y, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593(7860):586–590. doi:10.1038/s41586-021-03539-7
8. Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi:10.1016/j.tcb.2015.10.014
9. Kiziler AR, Aydemir B, Gulyasar T, Unal E, Gunes P. Relationships among iron, protein oxidation and lipid peroxidation levels in rats with alcohol-induced acute pancreatitis. Biol Trace Elem Res. 2008;124(2):135–143. doi:10.1007/s12011-008-8127-6
10. Sledzinski M, Borkowska A, Sielicka-Dudzin A, et al. Cerulein-induced acute pancreatitis is associated with c-Jun NH(2)-terminal kinase 1-dependent ferritin degradation and iron-dependent free radicals formation. Pancreas. 2013;42(7):1070–1077. doi:10.1097/MPA.0b013e318287d097
11. Ma X, Dong X, Xu Y, et al. Identification of AP-1 as a critical regulator of glutathione peroxidase 4 (GPX4) transcriptional suppression and acinar cell ferroptosis in acute pancreatitis. Antioxidants. 2022;12(1):100. doi:10.3390/antiox12010100
12. Yang L, Ye F, Liu J, Klionsky DJ, Tang D, Kang R. Extracellular SQSTM1 exacerbates acute pancreatitis by activating autophagy-dependent ferroptosis. Autophagy. 2023;19(6):1733–1744. doi:10.1080/15548627.2022.2152209
13. Li H, Lin Y, Zhang L, Zhao J, Li P. Ferroptosis and its emerging roles in acute pancreatitis. Chin Med J. 2022;135(17):2026–2034. doi:10.1097/CM9.0000000000002096
14. Shan Y, Li J, Zhu A, Kong W, Ying R, Zhu W. Ginsenoside Rg3 ameliorates acute pancreatitis by activating the NRF2/HO‑1‑mediated ferroptosis pathway. Int J Mol Med. 2022;50(1). doi:10.3892/ijmm.2022.5144
15. Jin H, Zhao K, Li J, Xu Z, Liao S, Sun S. Matrine alleviates oxidative stress and ferroptosis in severe acute pancreatitis-induced acute lung injury by activating the UCP2/SIRT3/PGC1alpha pathway. Int Immunopharmacol. 2023;117:109981. doi:10.1016/j.intimp.2023.109981
16. Ge P, Luo Y, Yang Q, et al. Ferroptosis in rat lung tissue during severe acute pancreatitis-associated acute lung injury: protection of Qingyi decoction. Oxid Med Cell Longev. 2023;2023:5827613. doi:10.1155/2023/5827613
17. Ma D, Li C, Jiang P, Jiang Y, Wang J, Zhang D. Inhibition of ferroptosis attenuates acute kidney injury in rats with severe acute pancreatitis. Dig Dis Sci. 2021;66(2):483–492. doi:10.1007/s10620-020-06225-2
18. Ma D, Jiang P, Jiang Y, Li H, Zhang D. Effects of lipid peroxidation-mediated ferroptosis on severe acute pancreatitis-induced intestinal barrier injury and bacterial translocation. Oxid Med Cell Longev. 2021;2021:6644576. doi:10.1155/2021/6644576
19. Kusske AM, Rongione AJ, Ashley SW, McFadden DW, Reber HA. Interleukin-10 prevents death in lethal necrotizing pancreatitis in mice. Surgery. 1996;120(2):284–288. doi:10.1016/S0039-6060(96)80299-6
20. Ji L, Guo X, Lv J, et al. Hypoxia-inducible factor-1alpha knockdown plus glutamine supplementation attenuates the predominance of necrosis over apoptosis by relieving cellular energy stress in acute pancreatitis. Oxid Med Cell Longev. 2019;2019:4363672. doi:10.1155/2019/4363672
21. Fang Z, Li J, Cao F, Li F. Integration of scRNA-Seq and bulk RNA-Seq reveals molecular characterization of the immune microenvironment in acute pancreatitis. Biomolecules. 2022;13(1):78. doi:10.3390/biom13010078
22. Kang R, Lotze MT, Zeh HJ, Billiar TR, Tang D. Cell death and DAMPs in acute pancreatitis. Mol Med. 2014;20(1):466–477. doi:10.2119/molmed.2014.00117
23. Su LJ, Zhang JH, Gomez H, et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longev. 2019;2019:5080843. doi:10.1155/2019/5080843
24. Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem Biol. 2020;27(4):420–435. doi:10.1016/j.chembiol.2020.02.005
25. Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100. doi:10.1016/j.semcancer.2019.03.002
26. Hou W, Xie Y, Song X, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–1428. doi:10.1080/15548627.2016.1187366
27. Liu J, Guo ZN, Yan XL, et al. Crosstalk between autophagy and ferroptosis and its putative role in ischemic stroke. Front Cell Neurosci. 2020;14:577403. doi:10.3389/fncel.2020.577403
28. Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. doi:10.15252/embj.2020104705
29. Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7(3):301–303. doi:10.4161/auto.7.3.14509
30. Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111(9):1208–1221. doi:10.1161/CIRCRESAHA.112.265819
31. Li J, Jia YC, Ding YX, Bai J, Cao F, Li F. The crosstalk between ferroptosis and mitochondrial dynamic regulatory networks. Int J Biol Sci. 2023;19(9):2756–2771. doi:10.7150/ijbs.83348
32. Lesina M, Wormann SM, Neuhofer P, Song L, Algul H. Interleukin-6 in inflammatory and malignant diseases of the pancreas. Semin Immunol. 2014;26(1):80–87. doi:10.1016/j.smim.2014.01.002
33. Yang J, Tang X, Ke X, Dai Y, Shi J. Triptolide suppresses NF-kappaB-mediated inflammatory responses and activates expression of Nrf2-mediated antioxidant genes to alleviate caerulein-induced acute pancreatitis. Int J Mol Sci. 2022;23(3):1.
34. Kong L, Deng J, Zhou X, et al. Sitagliptin activates the p62-Keap1-Nrf2 signalling pathway to alleviate oxidative stress and excessive autophagy in severe acute pancreatitis-related acute lung injury. Cell Death Dis. 2021;12(10):928. doi:10.1038/s41419-021-04227-0
35. Ma Y, Li X, Liu Z, Xue X, Wang Y, Ma Y. HIF-1alpha-PPARgamma-mTORC1 signaling pathway-mediated autophagy induces inflammatory response in pancreatic cells in rats with hyperlipidemic acute pancreatitis. Mol Biol Rep. 2023;50:8497–8507. doi:10.1007/s11033-023-08639-3
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.