Back to Journals » Breast Cancer: Targets and Therapy » Volume 9
An exploratory study of host polymorphisms in genes that clinically characterize breast cancer tumors and pretreatment cognitive performance in breast cancer survivors
Authors Koleck TA, Bender CM, Clark BZ, Ryan CM, Ghotkar P, Brufsky A , McAuliffe PF, Rastogi P, Sereika SM, Conley YP
Received 4 October 2016
Accepted for publication 8 December 2016
Published 3 March 2017 Volume 2017:9 Pages 95—110
DOI https://doi.org/10.2147/BCTT.S123785
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Theresa A Koleck,1,2 Catherine M Bender,1 Beth Z Clark,3,4 Christopher M Ryan,5,6 Puja Ghotkar,1 Adam Brufsky,4,7,8 Priscilla F McAuliffe,4,8,9 Priya Rastogi,4,7 Susan M Sereika,1,10,11 Yvette P Conley,1,12
1School of Nursing, University of Pittsburgh, Pittsburgh, PA, 2School of Nursing, Columbia University, New York, NY, 3Division of Gynecologic Pathology, Magee-Womens Hospital of University of Pittsburgh Medical Center (UPMC), 4School of Medicine, 5Department of Psychiatry, University of Pittsburgh, Pittsburgh, PA, 6Department of Psychiatry, University of California San Francisco, San Francisco, CA, 7Division of Hematology/Oncology, Magee-Womens Hospital of UPMC, 8University of Pittsburgh Cancer Institute, 9Division of Breast Surgical Oncology, Magee-Womens Hospital of UPMC, 10Department of Biostatistics, 11Department of Epidemiology, 12Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA
Purpose: Inspired by the hypothesis that heterogeneity in the biology of breast cancers at the cellular level may account for cognitive dysfunction symptom variability in survivors, the current study explored relationships between host single-nucleotide polymorphisms (SNPs) in 25 breast cancer-related candidate genes (AURKA, BAG1, BCL2, BIRC5, CCNB1, CD68, CENPA, CMC2, CTSL2, DIAPH3, ERBB2, ESR1, GRB7, GSTM1, MELK, MKI67, MMP11, MYBL2, NDC80, ORC6, PGR, RACGAP1, RFC4, RRM2, and SCUBE2), identified from clinically relevant prognostic multigene-expression profiles for breast cancer, and pretreatment cognitive performance.
Patients and methods: The sample (n=220) was comprised of 138 postmenopausal women newly diagnosed with early stage breast cancer and 82 postmenopausal age- and education-matched healthy controls without breast cancer. Cognitive performance was assessed after primary surgery but prior to initiation of adjuvant chemotherapy and/or hormonal therapy using a comprehensive battery of neuropsychological tests encompassing eight cognitive function composite domains: attention, concentration, executive function, mental flexibility, psychomotor speed, verbal memory, visual memory, and visual working memory. In total, 131 SNPs were included in the analysis. Standard and robust multiple linear regression modeling was used to examine relationships between each domain and the presence or absence of one or more minor alleles for each SNP. Genetic risk/protection scores (GRSs) were calculated for each domain to evaluate the collective effect of possession of multiple risk/protective alleles.
Results: With the exception of CMC2, MMP11, and RACGAP1, significant (P<0.05) SNP main effect and/or SNP by future prescribed treatment group interactions were observed for every gene between at least one domain and one or more SNPs. All GRSs were found to be significantly (P<0.001) associated with each respective domain score.
Conclusion: Associations between host SNPs and computed GRSs and variability in pretreatment cognitive function performance support the study hypothesis, and warrant further investigations to identify biomarkers for breast cancer-related cognitive dysfunction.
Keywords: breast neoplasms, genetics, cognition, biomarkers
Introduction
The recently published American Cancer Society/American Society of Clinical Oncology Breast Cancer Survivorship Care Guideline includes “assessment and management of physical and psychosocial long-term and late effects of breast cancer (BC) and treatment” as one of the five key areas of BC survivorship.1 Cognitive impairment related to cancer and cancer treatments is included in the guideline as a common and detrimental symptom experienced by BC survivors that can result in “distress and impaired [quality of life]”.1 While assessment and management of cognitive dysfunction in BC survivors by clinicians are recommended, the guideline acknowledges that the causes of and treatment for cognitive dysfunction are not well established.1 The guideline does not include recommendations for clinicians on how to predict which survivors will experience cognitive difficulties, the severity of the difficulties, or the duration of the impairment either.
The lack of biomarkers available to enhance precision survivorship care is in stark contrast to those that have been developed to refine outcome prediction and selection of optimal therapy for BC. Specifically, the introduction of advanced genetic technologies into patient care has greatly enriched the cellular-level characterization of breast neoplasms and led to the development of clinically relevant prognostic multigene-expression profiles for BC. Briefly, prognostic multigene-expression profiles for BC use tumor gene-expression algorithm-driven estimation to enrich prediction of long-term BC outcomes, including recurrence or metastasis, and/or benefit of adjuvant therapies.
Considering that many investigators theorize that cognitive difficulties, especially prior to adjuvant chemotherapy and/or hormonal therapy, are related to the cancer itself,2–5 we propose the use of BC-related genetic biomarkers for cognitive dysfunction symptom prediction and hypothesize that heterogeneity in BCs at the cellular level may account for variability in cognitive performance within the context of BC. Because genes utilized in multigene-expression profiles for BC contribute to characterizing BCs at the cellular level in relation to aggressiveness and risk of progression, they represent ideal candidate genes for a biomarker study to test our hypothesis.
The potential use of BC-related genetic markers to account for cognitive difficulties among BC survivors is not without evidence. A growing number of studies are investigating associations between host genetic variability and alterations in cognitive performance in women diagnosed with and receiving treatment for BC. Four published investigations have reported relationships between APOE and cognitive performance in women with BC.6–9 Associations with polymorphisms in genes involved in the dopamine and serotonin (ANKK1, BDNF, COMT, MTHFR, and SLC64A)9–11 and DNA repair and oxidative stress (CAT, ERCC2, ERCC3, ERCC5, GPX1, PARP1, SEPP1, SOD1, and SOD2)12 pathways have also been reported. However, to the best of our knowledge, no previous investigations have focused on BC-related genes as potential biomarkers for cognitive performance in women with BC.
To summarize, based on our hypothesis that heterogeneity in BCs at the cellular level may account for variability in cognitive performance within the context of BC, this study was conducted to explore the contribution of host polymorphisms within candidate genes and their regulatory regions known to differentiate BC heterogeneity at the cellular level to pretreatment (ie, postsurgery, preadjuvant therapy) cognitive performance in postmenopausal women diagnosed with BC.
Patients and methods
Participants
The sample (n=220) for this exploratory, genetic-association study was comprised of 138 postmenopausal women newly diagnosed with stage 1, 2, or 3A BC with no evidence of metastases and 82 postmenopausal age- and education-matched healthy controls (HCs) without BC. Participants were initially enrolled in a study examining the effects of the adjuvant antiestrogen therapy, anastrozole ± chemotherapy on cognitive function in postmenopausal women diagnosed with BC prior to, throughout, and following the antiestrogen-therapy regimen.13 Women diagnosed with BC were recruited from the Comprehensive BC Program of the University of Pittsburgh Cancer Institute. HC participants were obtained via referral from participants diagnosed with BC, advertisements, and random-digit dialing through the University Center for Social and Urban Research. All study participants were 75 years of age or younger, able to speak and read English, and had completed a minimum of 8 years of education. Participants were excluded if they had a prior history of neurologic disease or cancer or had been hospitalized for psychiatric illness within the past 2 years. For this study, in order to account for the heterogeneity of BC tumors, women diagnosed with BC were further classified using prescribed future-treatment regimen as a surrogate for disease characteristics. Therefore, the analysis included two cohorts of women diagnosed with BC – those prescribed chemotherapy followed by anastrozole (prescribed C+A) (n=55) and those prescribed anastrozole only (prescribed AO) (n=83) – as well as a cohort of HC women (n=82). All participants provided written informed consent for study participation. Both the current genetic ancillary study and the parent study were approved by the University of Pittsburgh Institutional Review Board.
Candidate-gene selection
A total of 25 biologically plausible candidate genes that are theorized to characterize the biology of BC at the cellular level through utilization in prognostic multigene-expression profiles for BC were selected for investigation. Detailed rationale for selection and biological plausibility of candidate genes has been discussed previously.5 Prognostic multigene-expression profiles for BC use tumor gene-expression algorithm-driven estimation to enrich prediction of long-term cancer outcomes (ie, recurrence or metastasis) and/or benefit of adjuvant therapy. A number of multigene-expression profiles for BC have been developed and include: the eleven-gene expression signature (Breast Cancer IndexSM; Biotheranostics, San Diego, CA, USA),14 the 14-gene prognostic expression signature (described in Tutt et al),15 the 21-gene BC assay (Oncotype DX® Breast Cancer Assay; Genomic Health, Redwood City, CA, USA),16,17 the 50-gene BC prognostic gene-signature assay (Prosigna® Breast Cancer Prognostic Gene Signature Assay; NanoString® Technologies Inc, Seattle, WA, USA) based on the PAM50 Breast Cancer Intrinsic Classifier,18 and the 70-gene BC-recurrence assay (MammaPrint® 70-gene Breast Cancer Recurrence Assay; Agendia®, Irvine, CA, USA).19,20 While the profiles vary in the number of genes utilized, patient-eligibility criteria, and specific prognostic goal, genes included in these profiles play an important role in characterizing the biology of BC at the cellular level to address aggressiveness and risk of progression, and thus point to ideal candidates for an initial investigation of the study hypothesis.
A total of 21 of the 25 candidate genes (BAG1, BCL2, BIRC5, CCNB1, CENPA, CMC2, DIAPH3, ERBB2, ESR1, GRB7, MELK, MKI67, MMP11, MYBL2, NDC80, ORC6, PGR, RACGAP1, RFC4, RRM2, and SCUBE2) were prioritized for this investigation, based on duplication in two or more of the previously named multigene-expression profiles.12 Because the 21-gene BC assay is currently the most widely used profile in the US, the four remaining cancer genes used as part of this assay but not duplicated in another profile (AURKA, CD68, CTSL2, and GSTM1) were also prioritized.
Single-nucleotide polymorphism (SNP) selection
SNPs representing each candidate gene were selected. Functional or putatively functional (ie, known to influence expression levels, associated with BC, or associated with a cognitive phenotype) polymorphisms within or directly upstream of candidate genes were identified from the literature. When a functional polymorphism was not identified and/or did not fully represent the gene of interest, tagging SNPs were selected using the Phase III HapMap database. Because the profiles from which candidate genes were selected rely upon gene-expression data, evaluation of DNA variability was extended ±2,500 bps beyond the gene to capture the UTR5’ and UTR3’ regulatory regions. Initial criteria for selection of tagging SNPs were as follows: R2≥0.8, minor allele frequency (MAF) ≥0.2, and selected for Caucasian ancestry, which represented the majority of study participants. The MAF criterion was ultimately relaxed to identify tagging SNPs for CTSL2, GRB7, MELK, MMP11, and RACGAP1. In addition, select polymorphisms in MIR125A,21,22 CCDC170,23–27 and NFE2L228 were included to represent more fully ERBB2, ESR1, and GSTM1, respectively. In total, 163 functional and tagging SNPs were identified.
Genotype data collection and quality control
Samples (3 mL of whole blood or 2 mL of saliva) were obtained for genotyping. DNA was extracted from peripheral blood leukocytes using a simple salting-out procedure29 or from saliva following the protocol and reagents supplied with Oragene® DNA-collection kits.30 The iPlex® MassArray platform (Sequenom, San Diego, CA, USA) was used as the primary genotyping method for this study. SNPs not conducive to genotyping with the iPlex platform were genotyped using TaqMan® allelic discrimination with the ABI Prism 7000 Sequence Detection System (SDS) and SDS software version 1.2.3 (Thermo Fisher Scientific, Waltham, MA, USA) or using a restriction fragment-length polymorphism–polymerase chain reaction approach.
Negative controls were included with all analyses. Genotypes were double-called by individuals blinded to participant phenotypes, and discrepancies were addressed by reviewing raw data or regenotyping. Participant genotypes were classified for data analysis based on the presence or absence of the minor allele (MA) (homozygous wild type compared to the combination of heterozygotes and homozygous-variant genotypes).
SNPs with call rates less than 90% or MAFs of less than 0.05 were omitted. For SNPs not meeting the 90% call-rate threshold but deemed essential for inclusion in the study (due to functional consequence, location within a candidate gene, or lack of alternative SNPs available within a given gene), secondary genotyping approaches were attempted. Alternative SNPs in linkage disequilibrium were selected for essential SNPs in instances of multiple failed genotyping attempts and/or lack of availability of alternative genotyping methods. Each SNP was tested for Hardy–Weinberg equilibrium (HWE) using χ2 goodness-of-fit or Fisher’s exact tests to identify potential genotyping errors.
Pretreatment cognitive function evaluation
Cognitive performance was assessed using a comprehensive battery of neuropsychological tests encompassing eight cognitive function composite domains:
attention – Cambridge Neuropsychological Test Automated Battery (CANTAB) Rapid Visual Information Processing Test31
concentration – Digit Vigilance Test32
executive function – CANTAB Stockings of Cambridge31 and CANTAB Spatial Working Memory31
mental flexibility – Delis Kaplan Executive Function System Color–Word Interference Test33
psychomotor speed – Grooved Pegboard34 and Digit Symbol Substitution Test35
verbal memory – Rey Auditory Verbal Learning Test,36 Verbal Fluency Test, and Rivermead Story Test37
visual memory – CANTAB Paired Associates Learning31 and Rey Complex Figure Test38
visual working memory – CANTAB Stockings of Cambridge31 and Rey Complex Figure Test.38
Women with BC completed the battery after surgery, but before initiation of prescribed C+A or AO adjuvant-therapy regimens. HCs completed the same neuropsychological test battery. Specifics related to the battery, creation of composite cognitive function domains, and z-score calculation have been reported previously.13 Please note that more negative z-scores designate poorer performance. Age (in years), estimated verbal intelligence (National Adult Reading Test – revised),39 depressive symptoms (Beck Depression Inventory II),40 anxiety (Profile of Mood States Tension–Anxiety subscale),41 fatigue (Profile of Mood States Fatigue–Inertia subscale),41 and current pain at time of assessment (Brief Pain Inventory)42 were also recorded.
Statistical analysis
Stata version 14.1 (StataCorp, College Station, TX, USA) and Statistical Package for the Social Sciences (SPSS) versions 23 and 24 (IBM Corporation, Armonk, NY, USA) were used to perform statistical analyses. Descriptive statistics were computed. Standard and robust multiple linear regression modeling was used to examine relationships between each domain and the presence (ie, homozygous-variant genotype plus heterozygous genotype) or absence (ie, homozygous wild-type genotype) of one or more MAs for each SNP. Both main SNP effects only and SNP–prescribed treatment group-interaction effect-regression models were fitted. In all models, HCs served as the reference group for the two prescribed treatment groups (ie, prescribed C+A or prescribed AO). Similarly, the wild-type genotype served as the reference group for possession of one or more MAs. All models were adjusted for age, estimated intelligence, and levels of depressive symptoms, anxiety, fatigue, and pain, and prescribed treatment group. Underlying assumptions were assessed. To lessen the impact of potentially influential points and adjust for heteroscedasticity, robust regression (generated using Huber weighting and biweighting iterations) model estimated regression coefficients and significance levels are reported.
Genetic risk/protection scores (GRSs) for each domain were then calculated to explore the influence of possession of multiple significant (P<0.05) genotypes on domain scores, as previously described.12 SNP MAs that were significantly (P<0.05) negatively or positively associated with a domain by either SNP main effects and/or SNP–prescribed treatment interaction effects were used in GRS calculations. A weighted calculation method, in which unstandardized robust regression coefficients from the individual models were multiplied by 0 (absence) or 1 (presence), based on a participant’s genotype and prescribed treatment-group membership and then summed, was used to assign greater risk/protection to MAs with stronger associations. A lower GRS conveys greater genetic risk for poorer cognitive function, and a higher GRS conveys greater genetic protection. GRSs were added as the final predictor to standard and robust multiple linear regression models adjusted for age, estimated verbal intelligence, levels of depressive symptoms, anxiety, fatigue, pain, and prescribed treatment group. Only participants with all genetic data necessary for calculation of a GRS were included in the GRS analysis.
Results
Participant characteristics
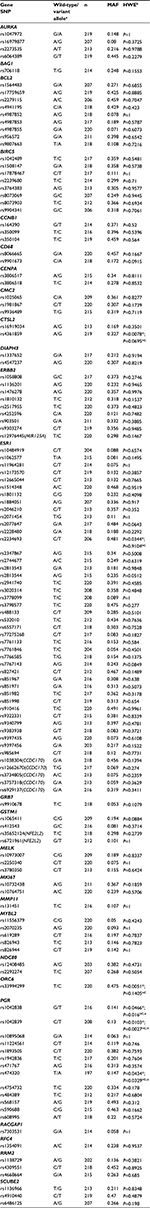
A total of 220 participants (n=55 prescribed C+A, n=83 prescribed AO, and n=82 HC) had genetic and complete covariate/confounder information and cognitive function scores available for one or more domains. A summary of overall demographic, covariate/confounder, and cognitive function data for participants included in this analysis can be found in Table 1.
Cohorts (ie, prescribed C+A, prescribed AO, and HC) differed statistically, yet not clinically meaningfully, by age and estimated verbal intelligence (Table 1). The groups also differed by level of anxiety (P=0.003), with women with BC prescribed C+A having higher mean pretreatment anxiety levels (9.61±6.14) than women with BC prescribed AO (6.97±4.654) and HCs (6.55±5.619). Comparison of tumor features by prescribed treatment group confirmed expected differences in disease characteristics (Table 2). To summarize, women with BC prescribed C+A had higher frequencies of American Joint Committee on Cancer Stage 2A, 2B, and 3A BCs, larger mean tumor size, higher mean number of positive lymph nodes, higher mean Nottingham Score, greater frequency of lymphovascular invasion, lower ER H-score, greater frequency of HER2-positive cancer, higher mean Ki67 index, and higher mean Oncotype DX® BC Assay Recurrence Score® compared to women with BC prescribed AO.
No differences in covariates/confounders or pretreatment cognitive function z-scores were observed between HCs included in this ancillary genetic analysis and those enrolled in the parent study but not included in the genetic analysis (n=82). Women with BC prescribed AO included in the genetic analysis did have slightly lower (P=0.044) mean estimated verbal intelligence (107.04±8.844) than those enrolled in the parent study but not included in the genetic analysis (n=155, 109.42±8.542). Also, women with BC prescribed C+A included in the genetic analysis had higher mean pretreatment verbal (P=0.014, 0.02±0.662), visual (P=0.006, 0.29±0.352), and visual working (P=0.002, 0.3±0.514) memory performance z-scores compared to those enrolled in the parent study but not included in the genetic analysis (n=78; –0.28±0.697, 0.03±0.615, –0.07±0.746, respectively).
Candidate-gene analysis
Of the 163 SNPs originally identified, 32 SNPs that were not amenable to multiplexing, had call rates less than 90%, or study MAFs of less than 0.05 were excluded. Alternatives were selected for three essential SNPs. In total, 131 SNPs were included in the genetic analysis (Table 3). Genotyping call rates for these SNPs ranged from 90% to 100%. When considering all study participants, six SNPs were not in HWE: CTSL2 rs4361859 (P=0.0078), ESR1 rs2234693 (P=0.0344), ORC6 rs33994299 (P=0.0051), PGR rs1042838 (P=0.0466), PGR rs1042839 (P=0.0103), and PGR rs474320 (P=0.0434). In HC women alone, PGR rs1042838 (P=0.016), PGR rs1042839 (P=0.0027), and PGR rs474320 (P=0.0329) still did not meet HWE. We attributed the deviations from HWE to nonrandom sampling of study participants from the population leading to enrichment for BC in the cases and de-enrichment for BC in the controls for these genes known to be involved in BC.
Individual polymorphisms significantly (P<0.05) associated with a domain by either SNP main effects or SNP–prescribed treatment group-interaction effects are summarized by domain in Table 4. Overall, significant relationships were noted between at least one domain and one or more polymorphisms of all candidate genes, except CMC2, MMP11, and RACGAP1. Comprehensive results from the individual SNP and cognitive function regression analyses are located in Table S1.
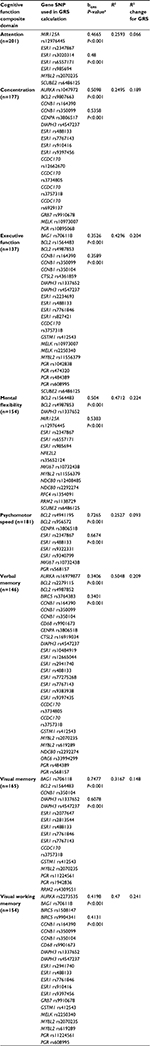
Therefore, one or more polymorphisms from the following genes, through either main SNP effects or SNP–prescribed treatment group-interaction effects, were included in GRSs: attention – ERBB2–MIR125A, ESR1, MYBL2, and SCUBE2; concentration – AURKA, BCL2, CCNB1, CENPA, DIAPH3, ESR1, ESR1–CCDC170, GRB7, MELK, and PGR; executive function – BAG1, BCL2, CCNB1, CTSL2, DIAPH3, ESR1, GSTM1, MELK, MYBL2, PGR, and SCUBE2; mental flexibility – BCL2, DIAPH3, ERBB2–MIR125A, ESR1, GSTM1–NFE2L2, MKI67, NDC80, RFC4, RRM2, and SCUBE2; psychomotor speed – BCL2, CENPA, ESR1, MKI67, and PGR; verbal memory – AURKA, BCL2, CCNB1, CD68, CENPA, CTSL2, DIAPH3, ESR1, ESR1–CCDC170, GSTM1, MYBL2, NDC80, ORC6, and PGR; visual memory – BAG1, BCL2, CCNB1, DIAPH3, ESR1, GSTM1, MYBL2, PGR, and RRM2; and visual working memory – AURKA, BAG1, BIRC5, CCNB1, CD68, DIAPH3, ESR1, GRB7, GSTM1, MELK, MYBL2, and PGR. All GRSs were found to be significantly (P<0.001) related to the respective domain score (Table 4). Reported associations were all positive, such that as GRS increased (ie, protection), cognitive function performance score improved (Figure 1).
Discussion
Individual candidate genes
In this first study exploring relationships among polymorphisms in biologically plausible BC-related candidate genes, we report significant relationships between performance on at least one cognitive function composite domain and one or more polymorphisms of all genes evaluated, with the exception of CMC2, MMP11, and RACGAP1. Significant findings related to the candidate genes found most broadly to impact cognitive function performance across multiple domains, specifically ESR1, CCDC170, PGR, CCNB1, MYBL2, BCL2, GSTM1, and DIAPH3, are discussed in detail in the following sections.
ESR1 and CCDC170
The ESR1 gene encodes an estrogen receptor. Polymorphisms in ESR1 have been previously associated with cognitive outcomes, including functioning, impairment, and Alzheimer’s disease.43 We found that performance on every cognitive domain was related to ESR1 polymorphisms through either main effects and/or interaction effects. The most global associations with a single ESR1 polymorphism occurred with an intronic upstream variant – rs488133. The effects of this polymorphism on cognitive function performance were different by domain and study cohort: rs488133-CT+TT contributed positively to executive function and psychomotor speed performance in all study participants. rs488133-CT+TT negatively impacted concentration performance in HCs, but positively impacted concentration performance in women with BC prescribed AO. In contrast, rs488133-CT+TT positively impacted memory performance in HCs, but negatively impacted memory performance in women with BC prescribed AO. In addition, while reported in other investigations of middle-aged and older women, we did not observe global cognitive impairment trends or memory deficits related to two well-studied polymorphisms in exon 1 of ESR1 named for the respective restriction enzyme-recognition sites: PvuII (rs2234693) and Xbal (rs9340799).44–47
Polymorphisms in CCDC170, the upstream neighbor of ESR1, were included in this study to represent more fully variability in ESR1. Associations between CCDC170 polymorphisms and BC susceptibility, progression, and survival have been reported.25,26,48–50 In addition, ESR1–CCDC170 chromosomal rearrangements have been associated with more aggressive estrogen receptor-positive BCs.51 While the function of CCDC170 is unknown, and no studies to date have investigated associations between CCDC170 polymorphisms and cognitive phenotypes, results from this analysis, in which possession of one or more CCDC170 MAs in four (rs12662670, rs3734805, rs3757318, and rs6929137) of the five SNPs evaluated was related to poorer concentration performance in all study participants, suggest that variation in CCDC170 plays an important role in concentration.
PGR
Progesterone receptors, encoded by PGR, are expressed throughout the brain in every neural cell type.52 Henderson et al found that progesterone concentrations were significantly and positively related to global cognition and verbal memory performance in healthy women less than 6 years since menopause.53 Moreover, Voytko et al found that estrogen plus progesterone improved executive function and attention performance in surgically menopausal monkeys.54 For executive function performance, we observed significant interactions between multiple PGR polymorphisms and study cohorts. In all instances, possession of PGR rs1042838–GT+TT, PGR rs474320–TA+AA, PGR rs484389–TC+CC, or PGR rs608995–AT+TT genotypes contributed positively to executive function-performance scores in HCs. When we looked at the interaction of these MAs within the context of BC, we saw the opposite effect: the combination of possession of one or more MAs and membership in a BC cohort was found to impact scores negatively, offsetting the positive SNP main effects and contributing an overall negative input to executive function performance in multiple instances.
The first SNP, rs1042838 (Val660Leu, G>T), is a missense polymorphism in exon 4 that is in linkage disequilibrium with rs1042839 (His770His, C>T), a silent polymorphism in exon 5, and a 320 bp Alu-element insertion at intron G; collectively, these polymorphisms form a variant haplotype called PROGINS. While the functional consequences remain unclear, the PROGINS allele has been associated with increased breast and ovarian cancer risk.55–59 Also evaluated in this study was rs474320, an intronic variant reported to be in tight linkage with PROGINS,60 and rs1042839, which is tightly linked to rs1042838. Both SNPs were found to be significant and as expected: rs1042839 generated very similar results to rs1042838; discrepancies in call rate may account for the differences in significance. The remaining significant SNPs, rs484389 and rs608995, are located in the UTR3’ of PGR. Taken together, these findings indicate that variation in regulation of progesterone receptors may be associated with executive function performance, and furthermore that the polymorphic impact on performance may vary in the systemic environment of a healthy individual compared to that of an individual diagnosed with BC.
CCNB1
CCNB1 encodes a cell-cycle regulatory protein important in mitosis.61 Because expression levels from this gene are used in three of five of the prognostic multigene-expression profiles for BC from which candidate genes were identified, CCNB1 was one of our top candidates for investigation of study hypotheses.5 Significant interactions were reported with study cohorts for three functional polymorphisms – rs164390 (102G>T), rs350099 (–957C>T), and rs350104 (–457C>T) – located in the promotor region of CCNB1 and memory and executive function performance. In general, we found that possession of rs164390GT+TT or rs350099CT+CC genotypes contributed positively to performance scores in HCs but close to zero or negatively in women with BC. The opposite contribution was observed for rs350104CT+CC genotypes. The genotypes associated with poorer cognitive performance in the cohorts of women with BC, rs164390–GT+TT, rs350099–CT+CC, and rs350104–TT, are all hypothesized to lead to lower levels of CCNB1 expression via reduced recruitment of transcription factors to the promotor region of the gene.62 This result is contradictory to anticipated findings, as higher cyclin B levels in breast tissue are associated with more severe cancer phenotypes.63,64 In addition, cyclin B levels were reported to be upregulated in autopsy hippocampal tissue in individuals with neuropathological Alzheimer’s disease and clinical dementia compared to individuals with normal aging.65 Nevertheless, the consistency of findings across three variants all theorized to impact expression in the same direction lends support to these associations. We would like to point out that one or more polymorphisms in the four other genes represented in three prognostic multigene-expression profiles for BC – CENPA, MELK, MYBL2, and ORC6 – were associated with performance on at least one domain.
MYBL2 and BCL2
MYBL2 encodes a nuclear protein, B-Myb, involved in cell-cycle progression and promotion of cell survival through activation of antiapoptotic genes.61,66 However, overexpression of B-Myb in certain settings induces apoptosis, and has been reported to contribute to neuronal cell death.66–69 We found significant relationships with two missense polymorphisms in MYBL2: rs11556379 (Ile624Met, C>G) and rs2070235 (Ser427Gly, A>G). The MAs of these polymorphisms have been reported to alter protein conformation, impair regulation of downstream targets, decrease antiapoptotic activity, and reduce cancer risk.70 Interestingly, for all study participants, rs2070235–AG+GG genotypes contributed positively to attention and negatively to memory-performance scores, while rs11556379–CG+GG genotypes contributed positively to mental flexibility-performance scores. We also reported a significant interaction related to executive function, where rs11556379–CG+GG genotypes had the opposite impact on performance in HCs (positive contribution to scores) and women with BC (negative contribution to scores).
Additionally, we report associations between polymorphisms in a gene regulated by MYBL2 that is also involved in apoptosis, BCL2, and concentration, executive function, mental flexibility, psychomotor speed, verbal memory, and visual memory performance. BCL2 expression has been associated with prognostication of disease-free survival, overall survival, and recurrence in BC.71–78 Moreover, normal breast tissue from women with BC was reported to display higher levels of BCL2 expression than breast tissue from women with no evidence of cancer.79 In relation to neurologic phenotypes, polymorphisms in BCL2 have been found to impact outcomes after traumatic brain injury and have been associated with hippocampal volume.80,81
GSTM1
One of the functional polymorphisms located in the promoter region of GSTM1, rs412543(–498C>G), was found to be important for memory and executive function performance. GSTM1 encodes an enzyme with antioxidant properties that detoxifies electrophilic compounds, including carcinogens, drugs, and environmental toxins, throughout the body.61 By decreasing the binding capability of the transcription factor AP2 to the GSTM1-promoter region, the G allele has been reported to decrease GSTM1 transcription by 30%–40% compared to the C allele.82 Both decreased and enhanced (attributed to counterproductive depletion of glutathione) GSTM1 expression has been associated with increased BC risk.82–84 We found that rs412543–GG+CG and hypothesized decreased GSTM1 expression contributed negatively to executive function and memory performance in all study participants. However, we also found positive interaction effects between rs412543–GG+CG and BC cohort related to verbal and visual working memory. While the mechanism is unclear, the paradoxical quality of GSTM1 under- and overexpression combined with study results suggests that decreased or moderate GSTM1 expression may be beneficial to certain aspects of cognitive function in women with BC. Considering the detoxification properties of GSTM1, further evaluation of cognitive decline over time in women with BC receiving adjuvant chemotherapy and/or antiestrogen therapy is recommended.
DIAPH3
Variation in the two upstream intronic polymorphisms selected to represent DIAPH3, rs1337652 and rs4547237, were associated with performance for multiple domains as well. DIAPH3 is involved in actin remodeling and regulation of cell movement and adhesion.61 DIAPH3 downregulation and silencing has been associated with metastatic disease due to loss of normal gene function and acquisition of an amoeboid cancer-cell phenotype.85 Evidence also suggests that DIAPH3 is critical to brain development and is involved in cell migration, the formation of dendrites and axons, axon guidance, and synaptic activity.86
CMC2, MMP11, and RACGAP1
Three candidate genes were not significantly associated with pretreatment cognitive performance in this study. While genes with significant findings from our analysis are represented by multiple functional and/or tagging SNPs and are well described in the literature, it is notable that the three genes not found to be significant are less well represented in the literature and the HapMap database. Single SNPs, rs131451 and rs7303531, were included in the analysis for MMP11 and RACGAP1, respectively. Both SNPs are upstream variants. No associations have been reported between MMP11 or RACGAP1 and cognitive phenotypes in the literature. CMC2 is an even more poorly described and studied gene, with reported involvement in cytochrome C oxidase activity.87 Two upstream (rs1025065 and rs1981867) polymorphisms and one downstream (rs9936489) polymorphism were identified using the Phase III HapMap database based on National Center for Biotechnology Information gene location (Chr16: 80975802…81006897), as CMC2 is not a displayed gene in HapMap. We must be mindful that our analysis is limited to current information known about these genes and polymorphisms, and thus these genes cannot be ruled out as important to understanding cognitive function within the context of BC.
Genetic risk/protection scores
Because of the complexity of BC as a disease and cognitive function as a phenotype, we calculated weighted GRSs for each domain to evaluate the collective effect of possession of multiple risk or protective MAs of genes used to clinically evaluate the biology of BC. Every GRS was significantly (P<0.001) and positively associated with its respective domain. When the GRSs were added as predictors to regression models, including age, estimated verbal intelligence, levels of depressive symptoms, anxiety, fatigue, pain, and prescribed treatment group, the explained variance (R2) increased by 0.066 to 0.244 for each domain. This substantial increase in R2 speaks to both the importance of host variation in genes used to evaluate clinically the biology of BC to pretreatment cognitive performance and the use of multiple common variants, plus personal and environmental factors, to model a complex phenotype.
Limitations and future directions
Small sample sizes limited our ability to conduct genetic analyses by genotype, rather than by the presence or absence of one or more MAs; therefore, we were unable to evaluate gene-dosage effects. In addition, the sample was comprised of postmenopausal women with hormone receptor-positive, early stage BC who were primarily Caucasian; therefore, the generalizability of study findings to premenopausal women, hormone-negative, different-stage BCs, or more diverse patient populations is unknown. The number of statistical tests completed as part of this exploratory study and possible inflation of type I error should also be acknowledged; all reported results will need to be confirmed in future independent studies. Limitations related to the prioritization and inclusion of select candidate genes has been discussed previously.5
While biomarkers of host DNA and cognitive performance are advantageous for a number of reasons, including the stability and tissue nonspecificity of DNA polymorphisms, associations with gene-expression and protein levels should also be conducted, as some of the most prominent findings from this study were related to polymorphisms with known functional consequences or located in regulatory regions. We postulate that cognitive performance variability in women with BC may be at least partially driven by tumor-gene expression and corresponding protein levels. Longitudinal studies that include cognitive assessment prior to primary surgery would be ideal for evaluation of the effect of tumor-gene expression, as well as changes in gene expression due to tumor removal and treatment of primary and secondary cancer sites, on variability in cognitive performance. Significant relationships from tumor gene-expression studies are advantageous for different reasons; namely, they could directly expand the clinical utility of currently marketed prognostic multigene-expression profiles for BC. Future analyses should also investigate the effect of polymorphisms in genes used to clinically evaluate the biology of BC and tumor-expression levels on cognitive function throughout and following adjuvant chemotherapy and/or antiestrogen-therapy regimens.
Conclusion
In summary, the objective of this study was to explore the hypothesis that host variation in candidate genes involved in BC development and prognosis is associated with variability in the presence and/or severity of alterations in pretreatment cognitive performance among postmenopausal women diagnosed with early stage BC. Significant associations between host polymorphisms representing 25 candidate genes used to clinically evaluate the biology of BC and computed GRSs and variability in pretreatment cognitive function performance support this hypothesis and merit independent replication and further investigation into the identification of clinically relevant biomarkers for BC-related cognitive dysfunction.
Acknowledgments
This study was funded by the National Institute of Nursing Research Cognitive Function and Breast Cancer: Genomics and Disease Characteristics (F31NR014590) and Targeted Research and Academic Training of Nurses in Genomics (T32NR009759), National Cancer Institute Long Term Trajectory of Cognitive Function Related to Anastrozole Use in Women (R01CA107408), American Cancer Society Doctoral Degree Scholarship in Cancer Nursing (DSCN-14-076-01-SCN), Oncology Nursing Society Foundation, a Sigma Theta Tau International Eta Chapter Research Award, a Nightingale Awards of Pennsylvania PhD degree scholarship, and a University of Pittsburgh School of Nursing Ruth and Bill Finke PhD Student research award.
Disclosure
The authors report no conflicts of interest in this work.
References
Runowicz CD, Leach CR, Henry NL, et al. American Cancer Society/American Society of Clinical Oncology breast cancer survivorship care guideline. J Clin Oncol. 2016;34(6):611–635. | ||
Wefel JS, Kesler SR, Noll KR, Schagen SB. Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J Clin. 2015;65(2):123–138. | ||
Ahles TA, Root JC, Ryan EL. Cancer- and cancer treatment-associated cognitive change: an update on the state of the science. J Clin Oncol. 2012;30(30):3675–3686. | ||
Ahles TA, Saykin AJ. Candidate mechanisms for chemotherapy-induced cognitive changes. Nat Rev Cancer. 2007;7(3):192–201. | ||
Koleck TA, Conley YP. Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer (Dove Med Press). 2016;8:29–37. | ||
Koleck TA, Bender CM, Sereika SM, et al. Apolipoprotein E genotype and cognitive function in postmenopausal women with early-stage breast cancer. Oncol Nurs Forum. 2014;41(6):E313–E325. | ||
Ahles TA, Saykin AJ, Noll WW, et al. The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psychooncology. 2003;12(6):612–619. | ||
Ahles TA, Li Y, McDonald BC, et al. Longitudinal assessment of cognitive changes associated with adjuvant treatment for breast cancer: the impact of APOE and smoking. Psychooncology. 2014;23(12):1382–1390. | ||
Lengacher CA, Reich RR, Kip KE, et al. Moderating effects of genetic polymorphisms on improvements in cognitive impairment in breast cancer survivors participating in a 6-week mindfulness-based stress reduction program. Biol Res Nurs. 2015;17(4):393–404. | ||
Ng T, Teo SM, Yeo HL, et al. Brain-derived neurotrophic factor genetic polymorphism (rs6265) is protective against chemotherapy-associated cognitive impairment in patients with early-stage breast cancer. Neuro Oncol. 2016;18(2):244–251. | ||
Small BJ, Rawson KS, Walsh E, et al. Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer. 2011;117(7):1369–1376. | ||
Koleck TA, Bender CM, Sereika SM, et al. Polymorphisms in DNA repair and oxidative stress genes associated with pre-treatment cognitive function in breast cancer survivors: an exploratory study. Springerplus. 2016;5:422. | ||
Bender CM, Merriman JD, Gentry AL, et al. Patterns of change in cognitive function with anastrozole therapy. Cancer. 2015;121(15):2627–2636. | ||
Jerevall PL, Ma XJ, Li H, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104(11):1762–1769. | ||
Tutt A, Wang A, Rowland C, et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer. 2008;8:339. | ||
Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. | ||
Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. | ||
Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. | ||
van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. | ||
Buyse M, Loi S, van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. | ||
Lehmann TP, Korski K, Ibbs M, Zawierucha P, Grodecka-Gazdecka S, Jagodziński PP. rs12976445 Variant in the pri-miR-125a correlates with a lower level of hsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol Lett. 2013;5(2):569–573. | ||
Hu Y, Liu CM, Qi L, et al. Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 2011;8(5):861–872. | ||
Barzan D, Veldwijk MR, Herskind C, et al. Comparison of genetic variation of breast cancer susceptibility genes in Chinese and German populations. Eur J Hum Genet. 2013;21(11):1286–1292. | ||
Han J, Jiang T, Bai H, et al. Genetic variants of 6q25 and breast cancer susceptibility: a two-stage fine mapping study in a Chinese population. Breast Cancer Res Treat. 2011;129(3):901–907. | ||
Hein R, Maranian M, Hopper JL, et al. Comparison of 6q25 breast cancer hits from Asian and European genome wide association studies in the Breast Cancer Association Consortium (BCAC). PLoS One. 2012;7(8):e42380. | ||
Stevens KN, Vachon CM, Lee AM, et al. Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res. 2011;71(19):6240–6249. | ||
Fletcher O, Johnson N, Orr N, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–435. | ||
Yu B, Lin H, Yang L, et al. Genetic variation in the Nrf2 promoter associates with defective spermatogenesis in humans. J Mol Med. 2012;90(11):1333–1342. | ||
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. | ||
DNA Genotek. Laboratory protocol for manual purification of DNA from whole sample. 2015. Available from: http://www.dnagenotek.com/US/pdf/PD-PR-015.pdf. Accessed December 14, 2016. | ||
Robbins T, James M, Owen A, Sahakian B, McInnes L, Rabbitt P. Cambridge Neuropsychological Test Automated Battery (CANTAB): a factor analytic study of a large sample of normal elderly volunteers. Dementia. 1994;5(5):266–281. | ||
Lafayette Clinical Instruments Company. Lafayette Clinical Repeatable Neuropsychological Battery. Sagamore (IN): Lafayette Clinical Instruments Company; 1989. | ||
Delis DC, Kaplan E, Kramer JH. Delis-Kaplan (D-KEFS) Executive Function System, Examiners Manual. San Antonio: Psychological Corporation; 2001. | ||
Klove H. Clinical neuropsychology. Med Clin North Am. 1963;47:1647–1658. | ||
Wechsler D. The Wechsler Memory Scale: Revised Manual. San Antonio: Psychological Corporation; 1998. | ||
Rey A. L’examen psychologique dans les cas d’encephalopathie traumatique (les problems) [The psychological examination in cases of traumatic encephalopathy: problems]. Arch Psychol. 1941;28:215–285. French. | ||
Cockburn J, Smith PT. Correlates of everyday memory among residents of Part III homes. Br J Clin Psychol. 1993;32(Pt 1):75–77. | ||
Osterrieth PA. Le test de copie d’une figure complexe: contribution a l’etude de la perception et de la memoire [Test of copying a complex figure: contribution to the study of perception and memory]. Arch Psychol. 1944;30:206–356. French. | ||
Nelson H. The National Adult Reading Test (NART): Test Manual. Windsor, UK: NFER-Nelson; 1982. | ||
Beck AT, Steer RA, Brown GK. Beck Depression Inventory-II. San Antonio: Psychological Corporation; 1996. | ||
McNair D, Lorr M, Droppleman LF. EdITS Manual for the Profile of Mood States (POMS). San Diego: Educational and Industrial Testing Service; 1992. | ||
Cleeland CS. Measurement of pain by subjective report. In: Chapman CR, Loeser JD, editors. Advances in Pain Research and Therapy. Vol 12. New York: Raven Press; 1989:391–403. | ||
Sundermann EE, Maki PM, Bishop JR. A review of estrogen receptor alpha gene (ESR1) polymorphisms, mood, and cognition. Menopause. 2010;17(4):874–886. | ||
Yaffe K, Lindquist K, Sen S, et al. Estrogen receptor genotype and risk of cognitive impairment in elders: findings from the Health ABC study. Neurobiol Aging. 2009;30(4):607–614. | ||
Bousman CA, Szoeke C, Chen K, Dennerstein L, Henderson VW, Everall IP. Oestrogen alpha-receptor variant and two-year memory decline in midlife Australian women. Neuropsychobiology. 2012;66(4):259–265. | ||
Kravitz HM, Meyer PM, Seeman TE, Greendale GA, Sowers MR. Cognitive functioning and sex steroid hormone gene polymorphisms in women at midlife. Am J Med. 2006;119(9 Suppl 1):S94–S102. | ||
Yaffe K, Lui LY, Grady D, Stone K, Morin P. Estrogen receptor 1 polymorphisms and risk of cognitive impairment in older women. Biol Psychiatry. 2002;51(8):677–682. | ||
Yamamoto-Ibusuki M, Yamamoto Y, Fujiwara S, et al. C6ORF97-ESR1 breast cancer susceptibility locus: influence on progression and survival in breast cancer patients. Eur J Hum Genet. 2015;23(7):949–956. | ||
Hong Y, Chen XQ, Li JY, et al. Current evidence on the association between rs3757318 of C6orf97 and breast cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2014;15(19):8051–8055. | ||
Fletcher O, Johnson N, Orr N, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–435. | ||
Veeraraghavan J, Tan Y, Cao XX, et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of estrogen-receptor positive breast cancers. Nat Commun. 2015;5:4577. | ||
Brinton RD, Thompson RF, Foy MR, et al. Progesterone receptors: form and function in brain. Front Neuroendocrinol. 2008;29(2):313–339. | ||
Henderson VW, St John JA, Hodis HN, et al. Cognition, mood, and physiological concentrations of sex hormones in the early and late postmenopause. Proc Natl Acad Sci U S A. 2013;110(50):20290–20295. | ||
Voytko ML, Murray R, Higgs CJ. Executive function and attention are preserved in older surgically menopausal monkeys receiving estrogen or estrogen plus progesterone. J Neurosci. 2009;29(33):10362–10370. | ||
Stenzig J, Schweikert A, Piasecki A, Höppner G, Eschenhagen T, Rau T. Progesterone receptor variants associated with the PROGINS haplotype exhibit functional properties similar to those of wild-type progesterone receptor. Pharmacogenet Genomics. 2012;22(8):629–641. | ||
Agoulnik IU, Tong XW, Fischer DC, et al. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J Clin Endocrinol Metab. 2004;89(12):6340–6347. | ||
Rockwell LC, Rowe EJ, Arnson K, et al. Worldwide distribution of allelic variation at the progesterone receptor locus and the incidence of female reproductive cancers. Am J Hum Biol. 2012;24(1):42–51. | ||
Liu T, Chen L, Sun X, et al. Progesterone receptor PROGINS and +331G/A polymorphisms confer susceptibility to ovarian cancer: a meta-analysis based on 17 studies. Tumor Biol. 2014;35(3):2427–2436. | ||
Romano A, Delvoux B, Fischer DC, Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J Mol Endocrinol. 2007;38(1–2):331–350. | ||
Lee E, Hsu C, Haiman CA, et al. Genetic variation in the progesterone receptor gene and risk of endometrial cancer: a haplotype-based approach. Carcinogenesis. 2010;31(8):1392–1399. | ||
NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. Epub 2014 Nov 14. | ||
Silvestre-Roig C, Fernández P, Mansego ML, et al. Genetic variants in CCNB1 associated with differential gene transcription and risk of coronary in-stent restenosis. Circ Cardiovasc Genet. 2014;7(1):59–70. | ||
Winters ZE, Hunt NC, Bradburn MJ, et al. Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer: association with prognosis. Eur J Cancer. 2001;37:2405–2412. | ||
Kawamoto H, Koizumi H, Uchikoshi T. Expression of the G2-M checkpoint regulators cyclin B1 and Cdc2 in nonmalignant and malignant human breast lesions: immunocytochemical and quantitative image analyses. Am J Pathol. 1997;150(1):15–23. | ||
Silva AR, Santos AC, Farfel JM, et al. Repair of oxidative DNA damage, cell-cycle regulation and neuronal death may influence the clinical manifestation of Alzheimer’s disease. PLoS One. 2014;9(6):e99897. | ||
Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 2005;41(16):2479–2484. | ||
Lui DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19(6):719–732. | ||
Liu DX, Biswas SC, Greene LA. B-Myb and C-Myb play required roles in neuronal apoptosis evoked by nerve growth factor deprivation and DNA damage. J Neurosci. 2004;24(40):8720–8725. | ||
Iyirhiaro GO, Zhang Y, Estey C, et al. Regulation of ischemic neuronal death by E2F4-p130 protein complexes. J Biol Chem. 2014;289(26):18202–18213. | ||
Schwab R, Bussolari R, Corvetta D, et al. Isolation and functional assessment of common, polymorphic variants of the B-MYB proto-oncogene associated with a reduced cancer risk. Oncogene. 2008;27(20):2929–2933. | ||
Aleskandarany MA, Soria D, Green AR, et al. Markers of progression in early-stage invasive breast cancer: a predictive immunohistochemical panel algorithm for distant recurrence risk stratification. Breast Cancer Res Treat. 2015;151(2):325–333. | ||
Lyng MB, Lænkholm AV, Tan Q, et al. Gene expression signatures that predict outcome of tamoxifen-treated estrogen receptor-positive, high-risk, primary breast cancer patients: a DBCG study. PLoS One. 2013;8(1):e54078. | ||
Kerr DA, Wittliff JL. A five-gene model predicts clinical outcome in ER+/PR+, early-stage breast cancers treated with adjuvant tamoxifen. Horm Cancer. 2011;2(5):261–271. | ||
Abdel-Fatah TM, Powe DG, Ball G, et al. Proposal for a modified grading system based on mitotic index and Bcl2 provides objective determination of clinical outcome for patients with breast cancer. J Pathol. 2010;222(4):388–399. | ||
Dawson SJ, Makretsov N, Blows FM, et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103(5):668–675. | ||
Bremer TM, Jacquemier J, Charafe-Jauffret E, Viens P, Birnbaum D, Linke SP. Prognostic marker profile to assess risk in stage I-III hormone receptor-positive breast cancer patients. Int J Cancer. 2009;124(4):896–904. | ||
Callagy GM, Webber MJ, Pharoah PD, Caldas C. Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer. 2008;8:153. | ||
Linke SP, Bremer TM, Herold CD, Sauter, G, Diamond, C. A multimarker model to predict outcome in tamoxifen-treated breast cancer patients. Clin Cancer Res. 2006;12(4):1175–1183. | ||
Batchelder AJ, Gordon-Weeks AN, Walker RA. Altered expression of anti-apoptotic proteins in non-involved tissue from cancer-containing breasts. Breast Cancer Res Treat. 2009;114(1):63–69. | ||
Sloan CD, Shen L, West JD, et al. Genetic pathway-based hierarchical clustering analysis of older adults with cognitive complaints and amnestic mild cognitive impairment using clinical and neuroimaging phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(5):1060–1069. | ||
Hoh NZ, Wagner AK, Alexander SA, et al. BCL2 genotypes: functional and neurobehavioral outcomes after severe traumatic brain injury. J Neurotrauma. 2010;27(8):1413–1427. | ||
Yu KD, Di GH, Fan L, et al. A functional polymorphism in the promoter region of GSTM1 implies a complex role for GSTM1 in breast cancer. FASEB J. 2009;23(7):2274–2287. | ||
Roodi N, Dupont WD, Moore JH, Parl FF. Association of homozygous wild-type glutathione S-transferase M1 genotype with increased breast cancer risk. Cancer Res. 2004;64(4):1233–1236. | ||
Reed DJ. Glutathione: toxicological implications. Annu Rev Pharmacol Toxicol. 1990;30:603–631. | ||
Hager MH, Morley S, Bielenberg DR, et al. DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med. 2012;4(8):743–760. | ||
Vorstman JA, van Daalen E, Jalali GR, et al. A double hit implicates DIAPH3 as an autism risk gene. Mol Psychiatry. 2011;16(4):442–451. | ||
Horn D, Zhou W, Trevisson E, et al. The conserved mitochondrial twin Cx9C protein Cmc2 is a Cmc1 homologue essential for cytochrome C oxidase biogenesis. J Biol Chem. 2010;285(20):15088–15099. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.