Back to Journals » International Journal of General Medicine » Volume 14
Age- and Gender-Independent Association of XRCC1 Arg399Gln Polymorphism with Chronic Myeloid Leukemia
Authors Abdalhabib EK , Jackson DE , Alzahrani B, Elfaki E , Hamza A , Mohamed Elasbali A , Alanazi F, Algarni A , Khider Ibrahim I , Saboor M
Received 20 September 2021
Accepted for publication 4 November 2021
Published 16 November 2021 Volume 2021:14 Pages 8231—8236
DOI https://doi.org/10.2147/IJGM.S340283
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Ezeldine K Abdalhabib,1 Denise E Jackson,2 Badr Alzahrani,1 Elyasa Elfaki,1 Alneil Hamza,1 Abdelbaset Mohamed Elasbali,1 Fehaid Alanazi,1 Abdulrahman Algarni,3 Ibrahim Khider Ibrahim,4 Muhammad Saboor5,6
1Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, AlQurayyat, Jouf University, Sakaka, Saudi Arabia; 2Thrombosis and Vascular Diseases Laboratory, School of Health and Biomedical Sciences, RMIT University, Victoria, Australia; 3Department of Medical Laboratory Technology, College of Applied Medical Sciences, Northern Borders University, Arar, Saudi Arabia; 4Department of Hematology, Faculty of Medical Laboratory Sciences, Al Neelain University, Khartoum, Sudan; 5Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Jazan University, Jazan, Saudi Arabia; 6Medical Research Center (MRC), Jazan University, Jazan, Saudi Arabia
Correspondence: Muhammad Saboor
Department of Medical Laboratory Technology, Faculty of Applied Medical Science, Jazan University, Jazan, Saudi Arabia
Tel +966 54 495 9029
Email [email protected]
Ezeldine K Abdalhabib
Department of Clinical Laboratory Sciences, College of Applied Medical Sciences-AlQurayyat, Jouf University, Saudi Arabia
Tel +966 538949566
Email [email protected]
Purpose: DNA damage to hematopoietic progenitor cells is an essential factor for leukemia development as a failure of the host DNA repair system to fix errors in DNA. This study aimed to assess the association of XRCC1 gene polymorphisms including Arg194Trp, Arg399Gln, and Arg280His with the risk of development of CML in Sudanese population.
Patients and Methods: The present study was conducted on 186 newly diagnosed patients with CML, aged 19– 70 years (118 males and 68 females; mean age of 46.15± 13.91 years) and 186 normal healthy controls (123 males and 63 females; mean age of 44.94± 8.97 years). Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) assay was utilized to analyze the XRCC1 (Arg194Trp, Arg399Gln, and Arg280His) gene polymorphisms.
Results: The genotypic frequencies of Arg399Gln polymorphism in cases were 131 (70.4%) homozygous Arg/Arg, 46 (24.7%) homozygous Gln/Gln, and 9 (4.8%) heterozygous Arg/Gln as compared to the controls ie, 153 (82.3%), 73 (14.5%), and 6 (3.2%), respectively. The Arg399Gln variant genotypic frequencies significantly differed between the cases and controls (χ2 = 7.249, P = 0.027). By comparison, no statistically significant difference was observed in the variant genotype frequencies between the cases and controls in terms of Arg194Trp and Arg280His polymorphisms.
Conclusion: XRCC1 Arg399Gln gene polymorphism might have an important role in increasing the risk of chronic myeloid leukemia among Sudanese patients. Furthermore, all tested three polymorphisms showed no association of risk of the development of CML with age and gender.
Keywords: XCCR1, chronic myeloid leukemia, polymorphism
Introduction
Chronic myeloid leukemia (CML) is a member of myeloproliferative neoplasms characterized by the acquired reciprocal chromosomal translocation, t(9;22) (q34; q11) which occurs as a result of translocation of ABL1 (Abelson murine leukemia) gene from chromosome 9 and its fusion with the BCR (breakpoint cluster region) gene on chromosome 22.1 Fusion of ABL gene with BCR gene encodes an oncoprotein known as BCR-ABL1 protein. BCR-ABL1 oncoprotein possess increased tyrosine kinase activity that leads to the relentless proliferation of myeloid lineage. This oncoprotein also interacts with other signaling pathways (STAT, MYC, RAS, RAF and JUN kinase) that also precipitate excessive proliferation.1 In addition to the uncontrolled tyrosine kinase activity, the BCR-ABL fusion gene leads to the production of increased intracellular reactive oxygen species that has spontaneous detrimental effects on DNA.2 DNA damage to hematopoietic precursor cells is an essential factor of leukemia development as the failure of the host DNA repair system to fix errors in the DNA.3–5 Many mechanisms participate in protecting genetic materials, such as DNA repair pathways. Defects in the DNA repair system and protein function could be affected by a large number of single-nucleotide polymorphisms (SNPs) that may cause many malignancies, including hematological neoplasms.6–9 Due to their pivotal role in maintaining the integrity of genome, the genes related with DNA repair have been studied extensively.
X-ray repair cross-complementing group 1 (XRCC1) is one of the most important genes embroiled in several processes including base excision repair, single-strand break repair, mismatch repair, and repair of double-strand breaks that restores the DNA damages occur to due to metabolic synthesis of reactive oxygen species (ROS), alkylating agents or radiations.10,11 Common lesions in cellular DNA are repaired through the interactions of XRCC1 with other DNA repair proteins eg, DNA ligase 3, poly ADP-ribose polymerase, and DNA polymerase-β.12 The number of SNPs in the XRCC1 gene have been found to be more than 300.13 Among them, three genetic polymorphisms have been widely studied: Arg194Trp (C→T substitution in codon 194 of exon 6), Arg399Gln (G→A substitution in codon 399 of exon 10), and Arg280His (G→A substitution in codon 280 of exon 9).3,14
The effect of the XRCC1 gene in the development of CML have been studied in different populations. However, in the Sudanese population, data related with the association of XRCC1 gene polymorphisms with CML are unavailable. The main aim of this study was to appraise the association of XRCC1 Arg194Trp, Arg399Gln, and Arg280His gene polymorphisms with the risk of development of CML in Sudanese population.
Patients and Methods
This was cross-sectional case-control study, conducted on 186 newly diagnosed patients with CML before starting the treatment, aged 19–70 years (118 males and 68 females; mean age of 46.15±13.91 years) and 186 normal healthy controls (123 males and 63 females; mean age of 44.94±8.97 years). This study was executed after obtaining ethical approval from the Ethics Committee of Al-Neelain University, Khartoum, Sudan. Written informed consent was obtained from each study participant following the guidelines of the Declaration of Helsinki. Patients with CML were recruited from the Hematology Clinics, Radiation and Isotope Center in Khartoum from August 2018 – December 2020. All CML cases were positive for Philadelphia chromosome and BCR-ABL gene rearrangement without a previous history or diagnosis of other malignancies. The patients were diagnosed by hematologist/oncologists in accordance with the WHO standards. Controls were selected from the same geographical area to represent a similar age range and ethnic background of the patients, but they had no previous history or present evidence of malignancy. The history and clinical data of the studied subjects were registered using a structural questionnaire with the help of hemato-oncologist. EDTA anticoagulated venous blood samples (5 mL) were collected from all participants in accordance with the PCR protocol.
DNA Extraction
DNA was extracted from the peripheral nucleated blood cells using QIAamp® DNA Mini kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer’s instructions. Quantitative (DNA and protein) and qualitative (purity and ratio) analyses of the DNA were carried out using a gene quant device (Amersham Bioscience–Biochrome Ltd., Cambridge, England). All the samples in the form of aliquots were stored at −20 °C until analysis.
Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) assay was utilized to analyze the XRCC1 (Arg194Trp, Arg399Gln, and Arg280His) gene polymorphisms. PCR was conducted to amplify exon 10 of the XRCC1 gene. The predesigned gene-specific primers and PCR protocol described by Annamaneni et al14 and Deligezer et al were followed for the detection of XRCC1 SNPs.9
Statistical Analysis
Data were analyzed and computed with the statistical package for social sciences (SPSS) version 25 (SPSS for Windows, Chicago, IL, USA) and SNPstat software. For descriptive parameters (mean and frequencies) and inferential statistics (Student’s t-test to compute the significance of the difference between the mean values of continuous variables). The difference between the proportions and categorical variables were calculated through Pearson’s Chi-square test (X2). Binary logistic regression was carried out to analyze the odds ratio (OR) and 95% confidence interval (CI). SNPstat software was used to calculate allele and genotype frequencies. The Hardy-Weinberg equilibrium (HWE) was used to compare the observed and expected genotype frequencies. The haplotypes and their frequencies were determined by using Linkage disequilibrium (LD) of the three loci. For each employed test the level of confidence P<0.05 was considered as a cutoff significance value.
Results
Demographic data of cases (patients with CML) and control, including age, gender, and phases of CML are depicted in Table 1. The cases and controls were age–gender matched. Cases had mean age of 46.15±13.9 years while 72.6% of them were >46 years of age. While mean age of the controls was 44.9±8.97 years, and 68.3% of them were > 46 years. The male group constituted 63.4% and 66.1% of cases and controls, respectively. Among the cases, 157 (84.4%) were diagnosed in the chronic phase, 21 (11.3%) accelerated phase, and 8 (4.3%) were in the blast crisis phase.
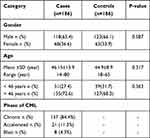 |
Table 1 Demographic Data of Patients with CML and Controls |
Among the cases, the most common genotype of Arg194Trp polymorphism was homozygous Arg/Arg 90.3% (n=168), while heterozygous Arg/Trp was found in 8.6% (n=16) and homozygous Trp/Trp in 1.1% (n=2) of the total populace. As for the controls, the genotype frequencies of homozygous Arg/Arg, heterozygous Agr/Trp and homozygous Trp/Trp were 171 (91.9%), 12 (6.5%), and 3 (1.6%), respectively. There was statistically insignificant difference in the variant genotype frequencies between the cases and controls (χ2 = 0.798, P = 0.671). Analysis of logistic regression suggested that the odds ratios for homozygous Arg/Arg were greater (OR = 1.474; 95% CI = 0.243–8.931) than those for heterozygous Arg/Trp (OR = 0.737; 95% CI = 0.338–1.604). The genotypic frequencies of Arg399Gln polymorphism in cases were found as; 131 (70.4%) homozygous Arg/Arg, 46 (24.7%) homozygous Gln/Gln, and 9 (4.8%) heterozygous Arg/Gln as compared to the controls ie, 153 (82.3%), 73 (14.5%), and 6 (3.2%), respectively. The variant genotype frequencies significantly differed between the cases and controls (χ2 = 7.249, P = 0.027). Odds ratios were greater for homozygous Gln/Gln (OR = 1.752; 95% CI = 0.608–5.052) than that for homozygous Arg/Arg (OR = 0.880; 95% CI = 0.282–2.745). Genotype and allele frequencies of XCCR1 Arg194Trp, Arg399Gln and Arg280His polymorphisms in the studied population are shown in Table 2.
 |
Table 2 Genotype of XCCR1 Arg194Trp, Arg399Gln and Arg280His Polymorphisms in the Studied Population |
Conversely, results of Arg280His polymorphism showed that a total of 174 (93.5%) cases were homozygous Arg/Arg, 1 (0.5%) homozygous His/His, and 11 (5.9%) heterozygous Arg/His and in controls, 173 (93%), 1 (0.5%), and 12 (6.5%), respectively. Statistically no significant difference was observed between the cases and controls (χ2 = 0.046, P = 0.977). Logistic regression suggested that the odds were slightly lower for homozygous His/His (OR = 1.006; 95% CI = 0.062–16.209) than those for heterozygous Arg/His (OR = 1.097; 95% CI = 0.471–2.554).
Allele frequencies of XCCR1 Arg194Trp (rs1799782), Arg399Gln (rs25487) and Arg280His (rs25489) polymorphisms in the studied population are shown in Table 3. For the Arg194Trp polymorphism, allele frequencies of Arg (C) and Trp (T) were nearly equal in cases as compared to controls (94.6%, and 5.4% vs 95.2%, and 4.8%). For the Arg399Gln polymorphism, the variant allele frequencies significantly differed between the cases and controls (P = 0.0003). The frequency of Arg (A) allele was 72.8% in cases and 83.9% in controls while the frequency of Gln allele (G) was 27.2% in cases whereas in controls it was 16.1%. Regarding the Arg280His polymorphism, the distribution of Arg (A) allele, and His (G) allele frequencies in cases and controls were nearly equal (96.5%, and 3.5% vs 96.2%, and 3.7%).
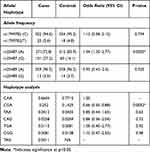 |
Table 3 The Alleles Frequency and Haplotypes Analysis Among Studied Subjects (N = 372) |
Table 4 illustrates the distribution of the studied polymorphisms and age of the patients at the time of diagnosis (age of more than mean age and those less than mean age). P-values were 0.972, 0.480 and 0.609 for Arg194Trp, Arg399Gln, and Arg280His polymorphisms, respectively. The studied polymorphisms also exhibited no impact on the risk of CML with respect to patients’ gender. Based upon gender distribution, the variant genotype frequencies of the patients with CML did not significantly differ (P-values were 0.782, 0.906, and 0.188) for Arg194Trp, Arg399Gln, and Arg280His polymorphisms, respectively, as shown in Table 4. Similarly, phase of CML did not demonstrate any association with Arg194Trp, Arg399Gln, and Arg280His polymorphisms, respectively, as depicted in Table 4.
 |
Table 4 Association of XCCR1 Gene Polymorphisms with Age, Gender and CML Phase |
Discussion
The genomic instability of the BCR-ABL expression in CML has the potential to create other mutations that cause DNA damage.15 The unrepaired cellular damage of DNA by exogenous and endogenous agents can lead to unregulated cell growth and neoplasia. DNA repair genes play a significant part in repairing this cellular damage and maintaining the genome integrity. XRCC1 has been studied in relation to different human cancers, including CML. Several studies have explored the association of CML and other types of leukemias in different population and ethnicities with the three most common XRCC1 gene polymorphisms, namely, Arg194Trp, Arg399Gln, and Arg280His.16–18 The present study is the first of its kind from Sudan to report XRCC1 gene polymorphisms in patients with CML. In this study, significant association of Arg399Gln polymorphism and CML was found as compared to healthy controls. The Arg399Gln, Gln399Gln, and Arg399Arg genotypes were detected in 70.4%, 24.7%, and 4.8% of the patients, respectively. This finding indicated the probable role of Arg399Gln polymorphism in CML in our population. These results agree with those of Annamaneni et al14 who found a correlation between the occurrence of polymorphisms in the XRCC1 gene and the progression of CML, but this correlation is inconsistent with the results of some other studies,9,19,20 who did not find any association between the XRCC1 codon 399Gln polymorphism and CML. A meta-analysis on XRCC1 Arg399Gln polymorphism and hematological malignancies reveals a positive association between the Arg399Gln polymorphism and an increase in the risk of developing hematological malignancies or leukemia in Asians.21
The presence of the Gln399 allele variant is accompanied with multiple abnormalities, including gene mutations, increased levels of sister chromatid exchanges, delay of the prolonged cell cycle, and considerably decreased DNA repair, as measured by the persistence of DNA adducts.22 These abnormalities may contribute to disease susceptibility in people with Arg399Gln polymorphism defects. The XRCC1 variant Arg280His allele has been reported to be associated with a reduction in the capacity of single-strand breaks and base excision repair system thus, this allele has been suggested as a potential risk factor of cancers.23
This study provided no evidence supporting the role of Arg280His polymorphism in susceptibility to CML in Sudanese patients. The distribution of this polymorphism was almost similar among CML cases and controls. However, this finding is contrary to some other reports with a significantly high risk of CML in individuals with Arg280His polymorphism.20,24 Additionally, Zhaodong et al25 conducted a meta-analysis and found that the XRCC1 gene in the Arg280His polymorphism is associated with leukemia susceptibility.
Arg194Trp polymorphism causes XRCC1 dysfunction via an amino acid (C→T) substitution in evolutionarily conserved regions, therefore, the risk of carcinogenesis increases.26 In the current study, we did not observe association of Arg194Trp polymorphism and increased risk of CML similar to the studies.20,24 However, this finding was in contrast to another study.27
This study did not find any correlation between the analyzed SNPs with the clinical phase of the disease, across the age groups of the study subjects upon diagnosis, and gender-related disease. However, findings about the association of XRCC1 Arg194Trp, Arg399Gln, and Arg280His polymorphisms, and the risk of CML were contradictory. Literature shows the age association with Arg/Gln more common in age group 20–40 years as compared to Gln/Gln homozygous which was noted in 40–60 aged group. In addition to that high frequency of Gln allele suggests an explicit role in the development of CML.14 The discrepancies between studies could be possibly due to study design, ethnicity of the studied populations, gene–environment interactions, life style, exposure to carcinogens, smoking and various roles of polymorphisms in different cell types or tissues that might have multiple effects on cancer formation.
Conclusion
In conclusion, XRCC1 Arg399Gln could be attributed as a risk factor of CML in Sudanese population irrespective age and gender.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this work through the grant number “375213500”. The authors would like to extend their sincere appreciation to the central laboratory at Jouf University for support this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2012 update on diagnosis, monitoring, and management. Am J Hematol. 2012;87(11):1037–1045. doi:10.1002/ajh.23282
2. Koptyra M, Falinski R, Nowicki MO, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108(1):319–327. doi:10.1182/blood-2005-07-2815
3. Bǎnescu C, Trifa AP, Demian S, et al. Polymorphism of XRCC1, XRCC3, and XPD genes and risk of chronic myeloid leukemia. Biomed Res Int. 2014;2014:213790. doi:10.1155/2014/213790
4. Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, Das-Gupta E. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100(10):3761–3766. doi:10.1182/blood-2002-04-1152
5. Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92(11):874–897. doi:10.1093/jnci/92.11.874
6. Batar B, Güven M, Bariş S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33(6):759–763. doi:10.1016/j.leukres.2008.11.005
7. Bolufer P, Barragan E, Collado M, Cervera J, López JA, Sanz MA. Influence of genetic polymorphisms on the risk of developing leukemia and on disease progression. Leuk Res. 2006;30(12):1471–1491. doi:10.1016/j.leukres.2006.01.016
8. Wang CQ, Krishnan V, Tay LS, et al. Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Rep. 2014;8(3):767–782. doi:10.1016/j.celrep.2014.06.046
9. Deligezer U, Akisik EE, Dalay N. Lack of association of XRCC1 codon 399Gln polymorphism with chronic myelogenous leukemia. Anticancer Res. 2007;27(4B):2453–2456.
10. Lévy N, Martz A, Bresson A, Spenlehauer C, de Murcia G, Ménissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34(1):32–41. doi:10.1093/nar/gkj409
11. Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279(53):55117–55126. doi:10.1074/jbc.M404524200
12. Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33(8):2512–2520. doi:10.1093/nar/gki543
13. Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58(4):604–608.
14. Annamaneni S, Gorre M, Kagita S, et al. Association of XRCC1 gene polymorphisms with chronic myeloid leukemia in the population of Andhra Pradesh, India. Hematology. 2013;18(3):163–168. doi:10.1179/1607845412Y.0000000040
15. Hehlmann R. How I treat CML blast crisis. Blood. 2012;120(4):737–747. doi:10.1182/blood-2012-03-380147
16. Duman N, Aktan M, Ozturk S, et al. Investigation of Arg399Gln and Arg194Trp polymorphisms of the XRCC1 (X-ray cross-complementing group 1) gene and its correlation to sister chromatid exchange frequency in patients with chronic lymphocytic leukemia. Genet Test Mol Biomarkers. 2012;16(4):287–291. doi:10.1089/gtmb.2011.0152
17. Tumer TB, Sahin G, Arinç E. Association between polymorphisms of EPHX1 and XRCC1 genes and the risk of childhood acute lymphoblastic leukemia. Arch Toxicol. 2012;86(3):431–439. doi:10.1007/s00204-011-0760-8
18. Mortada M, El-Sebaie A, Aladle D, Elzaafarany M, Mahmoud L. X-ray cross-complement 1 gene polymorphisms (Arg399Gln and Arg194Trp) in patients with acute myeloid leukemia. Egypt J Haematol. 2019;44(3):175. doi:10.4103/ejh.ejh_22_19
19. Ozdilli K, Pehlivan M, Serin I, Savran FO, Tomatir AG, Pehlivan S. DNA repair genes and chronic myeloid leukemia: ERCC2 (751), XRCC1 (399), XRCC4-intron 3, XRCC4 (−1394) gene polymorphisms. Mediterr J Hematol Infect Dis. 2021;13(1):3–7.
20. Bǎnescu C, Duicu C, Trifa AP, Dobreanu M. XRCC1 Arg194Trp and Arg399Gln polymorphisms are significantly associated with shorter survival in acute myeloid leukemia. Leuk Lymphoma. 2014;55(2):365–370. doi:10.3109/10428194.2013.802781
21. Yang X, Ma L, Zhao X, Yang H, Ruan L. A meta-analysis study on XRCC1 Arg399Gln polymorphism and hematological malignancies. Int J Clin Exp Med. 2016;9(10):19244–19255.
22. Geng J, Zhang YW, Huang GC, Chen LB. XRCC1 genetic polymorphism Arg399Gln and gastric cancer risk: a meta-analysis. World J Gastroenterol. 2008;14(43):6733–6737. doi:10.3748/wjg.14.6733
23. Takanami T, Nakamura J, Kubota Y, Horiuchi S. The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res. 2005;582(1–2):135–145. doi:10.1016/j.mrgentox.2005.01.007
24. Tang L, Xiong T, Jia Q, et al. Study on the association between the Arg194Trp polymorphism in the XRCC1 gene and the risk of hematological malignancies. Tumor Biol. 2014;35(4):3009–3016. doi:10.1007/s13277-013-1388-5
25. Zhang H, Liu H, Jiang G. Genetic polymorphisms of XRCC1 and leukemia risk: a meta-analysis of 19 case-control studies. PLoS One. 2013;8(11):e80687. doi:10.1371/journal.pone.0080687
26. Huang Y, Li L, Yu L. XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms in breast cancer risk: a meta-analysis. Mutagenesis. 2009;24(4):331–339. doi:10.1093/mutage/gep013
27. Du L, Liu Y, Xue P, et al. The Arg399Gln polymorphism in the XRCC1 gene is associated with increased risk of hematological malignancies. Tumor Biol. 2015;36(6):4545–4554. doi:10.1007/s13277-015-3099-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
