Back to Journals » The Application of Clinical Genetics » Volume 12
Adverse Drug Reactions Associated with CYP 2B6 Polymorphisms in HIV/AIDS-Treated Patients in Yaoundé, Cameroon
Authors Nguefeu Nkenfou C, Atogho Tiedeu B , Nguefeu Nkenfou C, Nji AM, Chedjou JP, Tah Fomboh C, Kouanfack C, Mbacham WF
Received 7 August 2019
Accepted for publication 23 November 2019
Published 30 December 2019 Volume 2019:12 Pages 261—268
DOI https://doi.org/10.2147/TACG.S226318
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Martin Maurer
Carine Nguefeu Nkenfou,1,2 Barbara Atogho Tiedeu,1,2 Celine Nguefeu Nkenfou,3–5 Akindeh M Nji,1,2 Jean Paul Chedjou,1,2 Calvino Tah Fomboh,2,6 Charles Kouanfack,7,8 Wilfred F Mbacham1,2,9
1Department of Biochemistry, Faculty of Science, University of Yaoundé I, Yaoundé, Cameroon; 2The Biotechnology Center, University of Yaoundé I, Yaoundé, Cameroon; 3Systems Biology, Chantal Biya’ International Reference Centre for Research on HIV and AIDS Prevention and Management (CBIRC), Yaoundé, Cameroon; 4Department of Biology, Higher Teachers’ Training College, University of Yaoundé I, Yaoundé, Cameroon; 5Molecular Biology Center Yaoundé, Yaoundé, Cameroon; 6Catholic University of Yaoundé (UCAC), Yaoundé, Cameroon; 7Day Care Unit, Central Hospital Yaoundé, Yaoundé, Cameroon; 8Department of Public Health, Faculty of Medicine and Pharmaceutical Sciences, University of Dschang, Dschang, Cameroon; 9Department of Public Health, Faculty of Medicine and Biomedical Sciences, University of Yaoundé I, Yaoundé, Cameroon
Correspondence: Wilfred F Mbacham
Laboratory for Public Health Research Biotechnologies, Biotechnology Center (BTC), P.O. Box 3851, Nkolbisson, Yaounde, Cameroon
Tel +237-677579180
Email [email protected]
Purpose: The metabolism of antiretroviral drugs is subject to individual variations of the CYP 2B6 gene. The objective of this study was to evaluate the prevalence of CYP 2B6 516 G>T and 983 T>C polymorphisms and investigate their association with the development of adverse drug reactions (ADRs) in people living with HIV/AIDS in Cameroon.
Patients and methods: A total number of 122 patients, attending the Yaoundé Central Hospital HIV Day Clinic, consented to take part in this study. Blood specimens were collected and DNA was extracted using the Chelex method. Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) was performed for the detection of CYP 2B6 Single-Nucleotide Polymorphisms (SNPs). Genotype frequencies were compared between groups with or without ADRs. Logistic regression analysis was performed to assess association between genotype and adverse effects of antiretroviral therapy (ART).
Results: Three types of metabolizers were identified: extensive, intermediate and slow. For the 516G>T polymorphism, prevalences of 8.2% GG, 65.6% GT and 26.2% TT were obtained. For the 983T>C polymorphism, 89.3% TT, 4.1% CT and 6.6% CC prevalences were obtained. Those homozygous for the wild-type allele (516GG) were less likely to develop ADR with a statistically significant difference (OR=0.885, P=0.029). For the CYP2B6 T983C SNP, homozygous mutants (CC) may present a higher risk (threefold) of developing adverse reactions (OR=2.677, P=0.164).
Conclusion: These findings demonstrate that ADRs among HIV/AIDS patients under ART may be associated with the genetic variability of the metabolizing enzyme CYP 2B6. Genotyping for this gene may guide the better administration of Efavirenz and Nevirapine to Cameroonian patients.
Keywords: CYP 2B6 polymorphisms, association, adverse drug reactions, HIV/AIDS
Introduction
Acquired Immunodeficiency Syndrome (AIDS) is one of the greatest public health challenges, with a World Health Organization (WHO) estimate of 36.7 million people infected around the world, and sub-Saharan Africa as the most affected region – accounting for 64% of the global burden.1 The disease is controlled by the administration of Antiretroviral Therapy (ART). The goals include the control of HIV replication; prevention of HIV transmission; reduction of HIV-related morbidity and mortality; and improving quality of life.2 The first line recommended by WHO in resource-limited countries, including Cameroon, consists of two Nucleoside Reverse Transcriptase Inhibitors (NRTI) plus one Non-Nucleoside Reverse Transcriptase Inhibitors (NNTIs) where Efavirenz (EFV) or Nevirapine (NVP) are the two most commonly used.3
Drug treatment in HIV disease is characterized by a great variability in response, in terms of both efficacy and toxicity.4 Several factors may affect this variability and may include ethnicity, gender, age, body weight, drug–drug and drug–food interactions, binding to plasma proteins, hepatic impairment, disease status, pregnancy, and host genetic factors.5 The benefit of pharmacogenetic testing is to guide the choice of the initial drug regimen, thus increasing efficacy, and simultaneously avoiding ADRs.6 Genetic variations can impact the pathways of drug absorption, disposition, metabolism and excretion (ADME).7 A mutation in a gene coding for a drug-metabolizing enzyme can result in an enzyme with normal, low, or no activity.8 EFV and NVP are principally metabolized by cytochrome P450 2B6.9,10
The gene that encodes for CYP2B6 is highly polymorphic.11 Up to date, about 60 allelic variants have been reported.12 Of these, CYP2B6 516G>T and 983T>C SNPs have being reported to be of clinical relevance.13 Several studies performed in Africa have reported that the CYP2B6 516G>T allele can occur in 20 to over 49% of the individuals.14–18 The second polymorphism is more frequent among African subjects with allele frequencies of 4–11%.13,19 These two polymorphisms have been associated with increased EFV and NVP plasma levels in several studies.20–28 A number of associations between these human genetic variants, high drug level and predisposition to ARV drug toxicity have been described in recent years.29–33 ADRs associated with NVP are cutaneous or dermatological events (toxidermia/hypersensitivity, skin rash, and pruritus). ADRs associated with EFV are central nervous events including insomnia, hallucinations, nightmares, headache, dizziness, and somnolence.
The objective of this work was to determine the frequency of CYP2B6 polymorphisms (516G>T and 983T>C) and the influence of their heterozygosity and homozygosity on the development of ADRs.
Materials and Methods
Study Setting, Design and Enrollment Procedure
The study was conducted in the Outpatients ART Centre of the Yaoundé Central Hospital (YCH), which is one of the largest in Cameroon. This unit was created in 1988 and follows about 10.000 HIV patients on ART treatment. The services provided include full consultation by devoted physicians (5) and psychosocial counselors (70), a pharmacy for drug refills, and laboratory testings (CD4 count, viral load and others) for biological follow-up. It is open from 8:00 a.m. to 3:30 p.m., and is one of the two Teaching Hospitals in Yaoundé, the other being the University Teaching Hospital.
HIV-infected individuals already under ART, with or without ADRs, were selected retrospectively, based on information reported in their medical records by clinicians after consultation. A list of patients with their telephone numbers was thus constituted. ADRs were diagnosed, based on patient complaints and/or physical changes noticed by physicians during routine clinical exam. ADR is said to be associated with therapy when absent before treatment and present after treatment initiation. Controls were recruited from the same Health Center and were selected on the basis of the absence of ADR development during at least 2 years of treatment regimen containing NVP or EFV. Participants were prospectively recruited after contacting them through phone calls. A total of 35 patients who had developed ADRs associated with NVP or EFV and 87 controls were recruited.
- Inclusion criteria:
- Under ART for at least 2 years
- >18 years
- Signed informed consent
- Being followed up
- Exclusion criteria:
- Withdrawal from the study.
Data Collection Procedure
Demographic, clinical and therapeutic data were obtained from clinical records of HIV/AIDS patients under ART who gave their informed consent. Such data included: sex, age, weight, CD4 cell counts, hemoglobin level, ART regimen, treatment initiation date, complaints after treatment initiation, ADR onset time, treatment modification and information on treatment observance or adherence.
Sample Collection and DNA Extraction
Five (5) mL of venous blood were collected and used to prepare dried blood spots on filter paper that were stored until genomic analysis. DNA was extracted using the Chelex method as previously described.34 The final supernatant (DNA) was transferred into a fresh tube and stored in TE buffer at −20°C for further pharmacogenetic analyses.
Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP)
Two alleles – CYP2B6 516G>T (rs3745274) and CYP 2B6 983T>C (rs28399499) – were investigated using PCR-RFLP. The primer sequences used to amplify the genes were CYP 516 F: 5ʹ-GGT CTG CCC ATC TAT AAA C-3ʹ and CYP 516 R: 5ʹ-CTG ATT CTT CAC ATG TCT GCG-3ʹ, CYP 983 F: 5ʹ-AGG AAT CCA CCC ACC TCA AC-3ʹ and CYP 983 R: 5ʹ-GAT AAG GCA GGT GAA GCA ATC A-3ʹ, respectively. The amplification was done in a T3 thermal cycler (Biometra, UK). Each PCR cycle was performed in a total volume of 25 μL containing: nuclease-free water, 10× ThermoPol buffer, 10 mM dNTPs (200 μM of each deoxyribonucleotide), 20 pmol of primer, 5 U/μL of Taq polymerase and 3 ng of gDNA. For 516 G>T, after initial denaturation at 95°C for 10 min, 35 cycles of amplification were carried out with denaturation at 94°C for 30 s, annealing at 58°C for 30 s and extension at 72°C for 60 s, followed by a final extension at 72°C for 10 min and then conservation at 4–8°C for 48 h. For 983T>C, the conditions were different at the level of annealing (59°C for 25 s) and the total number of cycles 45. To confirm the presence of CYP 2B6 gene, the expected amplicon sizes of the PCR products (526bp and 759bp, respectively) were verified on 2% agarose gel electrophoresis. The RFLP reaction conditions for digestion with BSrI (for 516 G>T) and BSmAI (for 983 T>C) (New England Biolabs, USA) were set at 65°C and at 55°C, respectively, both for 16 h each. The products of the digestion reaction were separated on a 2% agarose gel stained with ethidium bromide. Polymorphisms were determined according to specific fragment patterns as follows:
(516 G/G): 268 bp, 236 bp, 22 bp
(516 G/T): 504 bp, 268 bp, 236 bp, 22 bp
(516 T/T): 504 bp, 22 bp
(983 T/T): 759 bp
(983 C/T): 759 bp, 637 bp, 122 bp
(983 C/C): 637 bp, 122 bp
Compliance with Ethical Standards
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. An ethical clearance was obtained from the Comité National d’Ethique de la Recherche pour la Santé Humaine (Nº 2014/12/670/CE/CNERSH/SP).
Informed Consent
A written informed consent was obtained from each participant included in the study.
Data Analyses
All data were entered into Excel files and analyzed using the Statistical Package for Social Sciences (SPSS) version 16.0 (SPSS Inc., USA) statistical software. Frequencies of the CYP2B6 516G>T and 983T>C genotypes in the study population were obtained by descriptive statistics. The Chi-square test was used to assess the association between ADRs and baseline characteristics. The association between the genotype/phenotype of the CYP2B6 516G>T and 983T>C and adverse effects of ART (NVP and EFV) in the study population was evaluated by binary logistic regression analysis. The odds ratios (ORs) at 95% confidence intervals (CIs) were also calculated and the cut-off for statistical significance was set at a p value of <0.05.
Results
Study Population Characteristics
Of the 122 participants recruited, 74 (61%) were females while 48 (39%) were males. The mean age was 37.5 years with an average weight of 61.52 kg. Average hemoglobin was 10.82 g/dl and the mean CD4 cell count Pre-ART was 185.52 cells/mm3 (Table 1). Table 2 depicts the distribution of ADRs according to sex, age, weight, CD4 and hemoglobin. There were no statistically significant differences among the groups.
 |
Table 1 Baseline Characteristics of the Study Population |
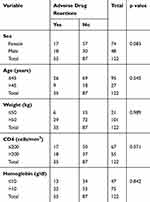 |
Table 2 Distribution and Association of Adverse Drug Reactions with Baseline Characteristics |
Adverse Reactions Reported Following ART Administration
For this study, we enrolled 87 patients with no ADRs (controls) and 35 with reported ADRs (cases). Among the 35 cases, 19 had developed cutaneous adverse events (toxidermia/hypersensitivity) and the rest (16) reported central nervous adverse events including insomnia, hallucinations, nightmares, headache, dizziness, and somnolence. The mean onset time (duration of patients on ART before developing reported ADR) was 2.69 (1–6) weeks. Additionally, in all of them, the treatment was modified due to these ADRs. Eight participants self-reported poor respect of treatment prescription.
Genotype and Allele Frequencies of CYP2B6 516G>T SNP Among Study Participants
The number of individuals presenting the CYP2B6 516GG wild-type genotype (extensive metabolizers) was 10 (8.2%). Seventy-nine (65.576%) participants had the heterozygous GT genotype (intermediate metabolizers) while 33 (26.23%) expressed the homozygous TT genotype considered as poor metabolizers (Figure 1). Allele frequencies for the CYP2B6 516G>T SNP were 40.96% for the G allele and 59.04% for the C allele.
 |
Figure 1 Genotype frequencies for the CYP2B6 516G>T SNP in the study population. Abbreviations: GG, Extensive Metabolizer; GT, Intermediate Metabolizer; TT, Slow metabolizer. |
Genotype and Allele Frequencies of CYP2B6 983 T>C SNP Among Study Participants
The number of individuals presenting the CYP2B6 983TT wild-type genotype (extensive metabolizers) was 109 (89.3%). Five (4.1%) had the heterozygous CT genotype (intermediate metabolizers) while eight (6.6%) expressed the homozygous CC genotype considered as poor metabolizers (Figure 2). Allele frequencies for the CYP2B6 983T>C SNP were 94.4% for T allele and 5.6% for C allele.
 |
Figure 2 Genotype frequencies for the CYP2B6 983T>C SNP in the study population. Abbreviations: TT, Extensive Metabolizer; CT, Intermediate Metabolizer; CC, Slow metabolizer. |
Association Between CYP2B6 516G>T SNP and CYP2B6 983T>C SNP Genotype/Phenotype of Participants and ADR
Association analysis of the CYP2B6 516G>T SNP showed that individuals homozygous for the wild-type allele were likely to have some degree of protection against ADRs with a statistically significant difference as compared to heterozygous and the mutants (OR=0.885, P=0.029). Additionally, the results showed that Individuals with heterozygous and homozygous mutants were at risk to develop ADRs (OR=1.208 and OR=1.34, respectively) although with no significant difference between them (P=0.416 and P=0.317, respectively). For the CYP2B6 T983C SNP, homozygote genotypes (CC) had a three times higher risk of developing adverse reactions (OR=2.677), but the difference with heterozygous and wild types was not statistically significant (P=0.164) (Table 3).
 |
Table 3 Association Between CYP2B6 516G>T SNP and CYP2B6 983T>C SNP Genotype/Phenotype of Participants with or Without ADR |
Haplotypes were constructed with the two variants explored in this study. The most frequent one was the 516GT/983TT and the 516GG/983CC was totally absent in the study population (Table 4).
 |
Table 4 Different Haplotypes and Their Prevalence in the Study Population |
Discussion
Data obtained on the distribution of genetic variations in populations are important in helping to understand inter-individual differences in response to treatment or drug. The association of the CYP 2B6 516 G>T and 983 T>C polymorphisms with Efavirenz and Nevirapine metabolism is well established. The frequencies of these SNPs were determined and their association with the development of adverse drug reactions to ART (NVP and EFV) investigated.
For the CYP 2B6 516 G>T polymorphism, individuals having the GG, GT and TT genotypes in our study population represented 8.2%, 65.6%, and 26.2%, respectively. The prevalence of TT in this study was lower than in West Africa (54%), Papua New Guinea (33%),35 Spain (40%)36 and Japan (32.6%).37 In Caucasians, Cambodians, and Burundese, considerably lower values of 5.9%, 9.2%, and 13%, respectively, were reported.21,38,39 Some studies in India reported 20.56–28% which are closer to our results.40,41 Huang et al reported a very low prevalence of the TT genotype (1.8%) among the Taiwanese.42 Regarding the other SNP 983 T>C, we obtained 89.3% TT, 4.1% CT and 6.6% CC. These results were not in line with that of Blievernicht and collaborators who found that this mutation was completely absent among individuals of white European origin.38
In our study, the minor allele frequencies for 516G>T and 983T>C were 0.41 (G) and 0.09 (C) respectively. These results were close to 0.34 and 0.07 obtained by Wyen et al on Germans,43 but a study on Guineans reported a very low frequency (0.01) of this C allele.19 The prevalence of the T allele (516G>T), which was observed in this study, was higher (59%) than the (35%) reported elsewhere (in Cambodia).39
It was observed that individuals homozygous for the wild-type allele (516 GG) were likely to be protected from ADR susceptibility with a statistically significant difference compared to heterozygous and mutants (OR=0.885, P=0.029). This could be due to the fact that extensive metabolizers eliminate drugs from the system rapidly; thus, such people are not exposed to high drug levels in the plasma. Our results equally showed that heterozygous and homozygous mutants for the 516G>T are at risk of developing ADRs due to the presence of the non-functional alleles (OR=1.208 and/or=1.34, respectively). For CYP2B6 SNP, T983C, participants with the homozygous genotypes (CC) were more susceptible (threefold higher risk OR=2.677) to develop conditions like cutaneous adverse reactions, central nervous adverse events regrouping insomnia, hallucination, nightmare, headache, dizziness, and somnolence, compared to those with other genotypes, although with no significance (P=0.164). This could be due to the fact that slow metabolizers eliminate drugs from the system slowly, thus leading to drug persistence resulting in toxicity. Our results can be compared to other studies that reported an association between the presence of 516G>T or 983T>C, high plasma NVP or EFV concentrations and the susceptibility to related ADRs.16,29–31,33,44 The lack of a significant difference between the presence of the adverse events and the genotypes might have been due to the small sample size, particularly the number of cases, which is one of the limitations of this study. The other limitation was the retrospective aspect of the participants’ selection. This study demonstrated that genetic variability in a metabolizing enzyme gene can also be correlated with susceptibility to ADR, a condition that should be considered. The fact that the association between ADR and both Efavirenz and Nevirapine was considered in this study is a strength because it was able to show that exposure to either of the drugs, your CYP 2B6 516G>T and 983 T>C genotype can create susceptibility to develop adverse effects. This is the first study carried out on HIV-treated patients in Cameroon that has considered associating the presence of polymorphisms on the CYP 2B6 gene with the susceptibility to develop ADRs.
Conclusion
This study was able to show that CYP2B6 gene variants (T983C and G516T) are associated with susceptibility to adverse drug reactions induced by Efavirenz or Nevirapine in HIV/AIDS-infected Cameroonians. It was found that the 983CC genotype confers a significantly higher risk of developing ADR. These findings need to be validated by a larger population. The results of this study suggest that CYP2B6 genotyping may help in optimizing antiretroviral therapy in patients who initiate an EFV- or NVP-based combination as it will help in identifying patients who will be more likely to develop adverse drug reactions in Cameroon.
Acknowledgments
The authors wish to acknowledge the contributions of the staff of the Yaoundé Central Hospital and all the study participants. The abstract of this paper was presented at the Cameroon Health Research Forum (CAHREF), held in Yaoundé–Cameroon as an oral presentation with interim findings: http://cahref.masante-cam.org/node/480.
Disclosure
The authors report no conflict of interest in this work.
References
1. UNAIDS (2017). Global Report: UNAIDSReport on the Global AIDS Epidemic 2017. Geneva: UNAIDS Guideline on When to Start Antiretroviral Therapy and on Pre-exposure Prophylaxis For HIV, World Health Organization; 2015. Available from: http://apps.who.int/iris/bitstream/10665/186275/1/9789241509565_eng.pdf?ua=1.
2. Cohen MS, Chen YQ, McCauley M, et al. Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med. 2016;375(9):830–839. doi:10.1056/NEJMoa1600693
3. Kouanfack C, Aghokeng AF, Mondain AM, et al. Lamivudine-resistant HBV infection in HIV-positive patients receiving antiretroviral therapy in a public routine clinic in Cameroon. Antivir Ther. 2011;17(2):321–326. doi:10.3851/IMP1911
4. Burger D, Heiden IVD, Porte CL, et al. Interpatient variability in the pharmacokinetics of the HIV non-nucleoside reverse transcriptase inhibitor efavirenz: the effect of gender, race, and CYP2B6 polymorphism. Br J Clin Pharmacol. 2006;61(2):148–154. doi:10.1111/j.1365-2125.2005.02536.x
5. Michaud V, Bar-Magen T, Turgeon J, et al. The dual role of pharmacogenetics in HIV treatment: mutations and polymorphisms regulating antiretroviral drug resistance and disposition. Pharmacol Rev. 2012;64(3):803–833. doi:10.1124/pr.111.005553
6. Bushyakanist A, Puangpetch A, Sukasem C, et al. The use of pharmacogenetics in clinical practice for the treatment of individuals with HIV infection in Thailand. Pharmgenomics Pers Med. 2015;8:163–170. doi:10.2147/PGPM.S86444
7. Barco AE, Novoa SR. The pharmacogenetics of HIV treatment: a practical clinical approach. J Pharmacogenomics Pharmacoproteomics. 2013;4:1. doi:10.4172/2153-0645.1000116
8. Banjoko B. Pharmacogenetics: the scientific basis. 2012. Available from: https://www.intechopen.com/books/pharmacology/pharmacogenetics-the-scientific-basis.
9. Erickson DA, Mather G, Trager WF, Levy RH, Keirns JJ. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab Dispos. 1999;27(12):1488–1495.
10. Ward BA, Jones DR, Stephen DH, et al. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306(1):287–300.
11. Kwara A, Lartey M, Sagoe KW, et al. CYP2B6 (c. dic516G→ T) and CYP2A6 (*9B and/or*17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67(4):427–436. doi:10.1186/2193-1801-1-34
12. Human cytochrome P450 (CYP) allele nomenclature database. Available from: http://www.cypalleles.ki.se/cyp2b6.htm.
13. Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet. 2013;4:1–12. doi:10.3389/fgene.2013.00024
14. Gross R, Aplenc R, Tenhave T, et al. Slow efavirenz metabolism genotype is common in Botswana. J Acquir Immune Defic Syndr. 2008;49(3):336–337. doi:10.1097/QAI.0b013e31817c1ed0
15. Nyakutira C, Röshammar D, Chigutsa E, et al. High prevalence of the CYP2B6 516G→T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol. 2008;64(4):357–365. doi:10.1007/s00228-007-0412-3
16. Mukonzo JK, Röshammar D, Waako P, et al. A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol. 2009;68(5):690–699. doi:10.1111/j.1365-2125.2009.03516.x
17. Brown KC, Hosseinipour MC, Hoskins JM, et al. Exploration of CYP450 and drug transporter genotypes and correlations with nevirapine exposure in Malawians. Pharmacogenomics. 2012;13(1):113–121. doi:10.2217/pgs.11.132
18. Sarfo FS, Zhang Y, Egan D, et al. Pharmacogenetic associations with plasma efavirenz concentrations and clinical correlates in a retrospective cohort of Ghanaian HIV-infected patients. J Antimicrob Chemother. 2014;69(2):491–499. doi:10.1093/jac/dkt372
19. Mehlotra RK, Bockarie MJ, Zimmerman PA. CYP2B6 983T>C polymorphism is prevalent in West Africa but absent in Papua New Guinea: implications for HIV/AIDS treatment. Br J Clin Pharmacol. 2007;64(3):391–395. doi:10.1111/j.1365-2125.2007.02884.x
20. Gozalo C, Gérard L, Loiseau P, et al. Pharmacogenetics of toxicity, plasma trough concentration and treatment outcome with nevirapine-containing regimen in anti-retroviral-naïve HIV-infected adults: an exploratory study of the TRIANON ANRS 081 trial. Basic Clin Pharmacol Toxicol. 2011;109(6):513–520. doi:10.1111/j.1742-7843.2011.00780.x
21. Calcagno A, D’Avolio A, Simiele M, et al. Influence of CYP2B6 and ABCB1 SNPs on nevirapine plasma concentrations in Burundese HIV-positive patients using dried sample spot devices: dried spots-based pharmacogenetics of nevirapine. Br J Clin Pharmacol. 2012;74(1):134–140. doi:10.1111/j.1365-2125.2012.04163.x
22. Holzinger ER, Grady B, Ritchie MD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS clinical trials group protocols implicates several CYP2B6 variants. Pharmacogenet Genomics. 2012;22(12):858–867. doi:10.1097/FPC.0b013e32835a450b
23. Sukasem C, Chamnanphon M, Koomdee N, et al. High plasma efavirenz concentration and CYP2B6 polymorphisms in Thai HIV-1 infections. Drug Metab Pharmacokinet. 2013;28(5):391–397. doi:10.2133/dmpk.DMPK-12-RG-120
24. Swart M, Skelton M, Ren Y, et al. High predictive value of CYP2B6 SNPs for steady-state plasma efavirenz levels in South African HIV/AIDS patients. Pharmacogenet Genomics. 2013;23(8):415–427. doi:10.1097/FPC.0b013e328363176f
25. Dickinson L, Chaponda M, Carr DF, et al. Population pharmacokinetic and pharmacogenetic analysis of nevirapine in hypersensitive and tolerant HIV-infected patients from Malawi. Antimicrob Agents Chemother. 2014;58(2):706–712. doi:10.1128/AAC.02069-13
26. Oluka MN, Okalebo FA, Guantai AN, et al. Cytochrome P450 2B6 genetic variants are associated with plasma nevirapine levels and clinical response in HIV-1 infected Kenyan women: a prospective cohort study. AIDS Res Ther. 2015;12:10. doi:10.1186/s12981-015-0052-0
27. Damronglerd P, Sukasem C, Thipmontree W, et al. A pharmacogenomic prospective randomized controlled trial of CYP2B6 polymorphisms and efavirenz dose adjustment among HIV-infected Thai patients: a pilot study. Pharmgenomics Pers Med. 2015;8:155–162. doi:10.2147/PGPM.S86446
28. Swart M, Evans J, Skelton M, et al. An expanded analysis of pharmacogenetics determinants of efavirenz response that includes 3′-UTR single nucleotide polymorphisms among Black South African HIV/AIDS patients. Front Genet. 2016;6:356. doi:10.3389/fgene.2015.00356
29. Gounden V, van Niekerk C, Snyman T, et al. Presence of the CYP2B6 516G> T polymorphism, increased plasma efavirenz concentrations and early neuropsychiatric side effects in South African HIV-infected patients. AIDS Res Ther. 2010;7(32):1–9. doi:10.1186/1742-6405-7-32
30. Yuan J, Guo S, Hall D, et al. Toxicogenomics of nevirapine-associated cutaneous and hepatic adverse events among populations of African, Asian, and European descent. AIDS Lond Engl. 2011;25(10):1271–1280. doi:10.1097/QAD.0b013e32834779df
31. Ciccacci C, Di Fusco D, Marazzi MC, et al. Association between CYP2B6 polymorphisms and nevirapine-induced SJS/TEN: a pharmacogenetics study. Eur J Clin Pharmacol. 2013;69(11):1909–1916. doi:10.1007/s00228-013-1549-x
32. Carr DF, Chaponda M, Cornejo Castro EM, et al. CYP2B6 c.983T>C polymorphism is associated with nevirapine hypersensitivity in Malawian and Ugandan HIV populations. J Antimicrob Chemother. 2014;69(12):3329–3334. doi:10.1093/jac/dku315
33. Gallien S, Journot V, Loriot MA, et al. Cytochrome 2B6 polymorphism and efavirenz-induced central nervous system symptoms: a substudy of the ANRS ALIZE trial. HIV Med. 2017;18(8):537–545. doi:10.1111/hiv.12488
34. Plowe CV, Djimde A, Bouare M, et al. Pyrimethamine and proguanil resistance-conferring mutations in plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52(6):565–568. doi:10.4269/ajtmh.1995.52.565
35. Mehlotra RK, Ziats MN, Bockarie MJ, et al. Prevalence of CYP2B6 alleles in malaria-endemic populations of West Africa and Papua New Guinea. Eur J Clin Pharmacol. 2006;62(4):267–275. doi:10.1007/s00228-005-0092-9
36. Rodriguez-Novoa S, Barreiro P, Rendon A, et al. Influence of 516G>T polymorphisms at the gene encoding the CYP450-2B6 isoenzyme on efavirenz plasma concentrations in HIV-infected subjects. Clin Infect Dis. 2005;40(9):1358–1361. doi:10.1086/429327
37. Gatanaga H, Hayashida T, Tsuchiya K, et al. Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6 *6 and *26. Clin Infect Dis. 2007;45:1230–1237. doi:10.1086/522175
38. Blievernicht J, Schaeffeler E, Klein K, et al. MALDI-TOF mass spectrometry for multiplex genotyping of CYP2B6 single-nucleotide polymorphisms. Clin Chem. 2007;53(1):24–33. doi:10.1373/clinchem.2006.074856
39. Chou M, Bertrand J, Segeral O, et al. Population pharmacokinetic-pharmacogenetic study of nevirapine in HIV-infected cambodian patients. Antimicrob Agents Chemother. 2010;54(10):4432–4439. doi:10.1128/AAC.00512-10
40. Ramachandran G, Hemanth AK, Rajasekaran S, et al. CYP2B6 G516T polymorphism but not rifampin coadministration influences steady-state pharmacokinetics of efavirenz in human immunodeficiency virus-infected patients in South India. Antimicrob Agents Chemother. 2009;53(3):863–868. doi:10.1128/AAC.00899-08
41. Varshney E, Saha N, Tandon M, Shrivastava V, Ali S. Prevalence of poor and rapid metabolizers of drugs metabolized by CYP2B6 in North Indian population residing in Indian national capital territory. SpringerPlus. 2012;1(1):34. doi:10.1186/2193-1801-1-34
42. Huang SH, Lin SW, Chang SY, et al. Prediction of plasma efavirenz concentrations among HIV-positive patients taking efavirenz-containing combination antiretroviral therapy. Sci Rep. 2017;7:1. doi:10.1038/s41598-017-16483-2
43. Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61(4):914–918. doi:10.1093/jac/dkn029
44. Ribaudo HJ, Liu H, Schwab M, et al. Impact of CYP2B6, ABCB1 and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS clinical trials group study. J Infect Dis. 2010;202:717–722. doi:10.1086/655470
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
