Back to Journals » Infection and Drug Resistance » Volume 16
Advance on Engineering of Bacteriophages by Synthetic Biology
Authors Sun Q, Shen L, Zhang BL, Yu J, Wei F, Sun Y, Chen W , Wang S
Received 12 January 2023
Accepted for publication 16 March 2023
Published 31 March 2023 Volume 2023:16 Pages 1941—1953
DOI https://doi.org/10.2147/IDR.S402962
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Qingqing Sun,1,* Lixin Shen,1,* Bai-Ling Zhang,2 Jiaoyang Yu,1,3 Fu Wei,1 Yanmei Sun,1 Wei Chen,3,4 Shiwei Wang1
1Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, Provincial Key Laboratory of Biotechnology of Shaanxi Province, the College of Life Sciences, Northwest University, Xi’an, 710069, People’s Republic of China; 2Department of Laboratory Medicine, The Second Affiliated Hospital of Nanchang University, Nanchang, 330006, People’s Republic of China; 3Clinical Research Center, the Second Hospital of Nanjing, Nanjing University of Chinese Medicine, Nanjing, 210003, People’s Republic of China; 4The Clinical Infectious Disease Center of Nanjing, Nanjing, 210003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Chen; Shiwei Wang, Email [email protected]; [email protected]
Abstract: Since bacteriophages (phages) were firstly reported at the beginning of the 20th century, the study on them experiences booming-fading-emerging with discovery and overuse of antibiotics. Although they are the hotspots for therapy of antibiotic-resistant strains nowadays, natural phage applications encounter some challenges such as limited host range and bacterial resistance to phages. Synthetic biology, one of the most dramatic directions in the recent 20-years study of microbiology, has generated numerous methods and tools and has contributed a lot to understanding phage evolution, engineering modification, and controlling phage-bacteria interactions. In order to better modify and apply phages by using synthetic biology techniques in the future, in this review, we comprehensively introduce various strategies on engineering or modification of phage genome and rebooting of recombinant phages, summarize the recent researches and potential directions of phage synthetic biology, and outline the current application of engineered phages in practice.
Keywords: phage, synthetic biology, engineering, rebooting, phage therapy, phage application
Introduction
Bacteriophage (phage) is a class of widespread viruses exclusively infecting bacteria, playing a key role in ecosystems.1–3 Since phages were discovered over a hundred years ago, many techniques and reagents, such as DNA polymerase, restriction endonuclease, ligase, and CRISPR/Cas system, have been developed from phages, which have together accelerated the development of modern biology. In addition, although the genetic and functional diversity of phages remains unclear, they provide an extremely rich library of genetic elements and toolkits for synthetic biology.4–7 From an application perspective, phages are believed to have great potential in antimicrobial agent, phage therapy, biosensor, and delivery vector.8,9 In particular, as a potential alternative to antibiotics, phages are considered as the white hope in solving the problem of the emergence and spread of antimicrobial resistance (AMR). So far, even though abundant phages have been isolated and characterized worldwide, many aspects about phage genes and life cycles are still mysterious, which limiting direct and extensive use of natural phages in clinical medicine.
Synthetic biology refers to the rational design, transformation, and even de novo synthesis of organisms according to specific goals under the guidance of engineering, which is a good way to overcome some limitations of natural bacteriophages.10–12 The efficiency of phage infection can be enhanced by adding functional genes to the phage genome. The virulence genes and genes with non-essential functions are removed as far as possible, and then the chassis genome of phage is used to achieve the purpose of highly controllable biosafety.13–15 New synthetic biology strategies and methods for bacterial genomes, such as high-throughput sequencing technology and large DNA fragment synthesis technology, can no doubt accelerate the ability of phage genome design and construction, and further, explore the potential of phage in all aspects. Advances about phage display are well summarized in other reviews.16,17 In this review, we focus on strategies on engineering or modification of phage genome and rebooting of recombinant phages, narrate and discuss the recent research and potential directions of phage synthetic biology, and summarize the current application of engineered phages in practice.
Engineering Strategies for Phage Genome
Genetically Engineered Phage
Although phages have application potential in many aspects, such as phage therapy, precision medicine, and bacterial prevention and control, native lytic phages remain an underutilized option due to challenges such as regulation, limited host range, bacterial resistance to phages, manufacturing, and side effects of bacterial lysis and delivery.18 These limitations can be potentially overcome by genetically engineered phages. In this section, we mainly discuss the kinds of means and methods used to modify lytic phages.
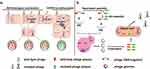
Currently, various methods of phage genetic engineer with advantages and disadvantages have been developed, in order to modify their host range, improve safety and antimicrobial activity.19 Homologous recombination (HR) in vivo is an early popular approach of phage genome editing, which allows gene insertion, replacement, and deletion (Figure 1a).20 By this means, linear dsDNA phages, such as Mycobacterium smegmatis phage L5 and Pseudomonas aeruginosa phage PaP1, can be modified.21,22 It is worthy to note that HR is feasible only when the host strains are competent for the donor DNA and restriction-deficient. However, the recombination efficiency of HR is always unsatisfactory, sometimes as low as 0.1%, requiring intensive labor to select recombinant phages. A selection marker inserted into the donor DNA is helpful.23,24 One example is trxA gene, which is essential for phage replication but not essential for host growth. The screening efficiency of T7 recombinants was higher in E. coli ∆trxA mutant compared with the wild-type E. coli strain.24,25 In addition, overexpression of heterologous recombinant proteins, such as λ red system (Exo, Gam and Beta) and RecE/RecT-like proteins in the host can improve the recombination efficiency.20,26,27 Mycobacteriophages Che9c gp60 and gp61 encode homologs of both RecE and RecT, which could substantially enhance mycobacterial recombination frequencies.20 For example, they facilitated the construction of M. smegmatis phage BPs gene knockout and replacement mutant, with efficiency up to 15%.28
CRISPR Cas9 system is widely used for gene knockout, insertion, and site-directed mutation of phage genome (Figure 1a).29–32 Martel lab confirmed that it can be used for mutation and large fragment deletions of S. thermophilus phage 2972 genome. They successfully achieved phage recombination, and all the tested plaques contained recombinant phages that had the desired mutation.33 In 2017, Aslan lab used natural CRISPR Cas10 system of S. epidermidis to engineer staphylococcal lytic phages Andhra, achieving silent mutation efficiency up to 100%.34 Besides, 11 kb to 29 kb of phage T4 genome was deleted by utilizing CRISPR Cas12a system, and all plaques were mutant.35 Of course, type I-E CRISPR Cas systems are also powerful tools for phage genome editing. With them, Vibrio cholerae phage ICP1_2011_A’s 33 bp gene deletion and 2670 bp gene deletion efficiency were 100% and 58%, respectively.36 Editing efficiency of CRISPER system is influenced by homologous-arm size of donor fragment, and the size of target sequence. For example, a 500 bp fragment in the Klebsiella bacteriophage phiKpS2 was replaced with 923 bp using homologous arms of 40 bp, 50 bp and 60 bp, the success rate was 41.7%, 60.4%, and 87.5%, respectively.31 An editing efficiency of 76.7% and 56.7% for deletion of a 1 kb and 2 kb fragment, respectively, could be obtained with a 40 bp homologous arm.37 Taken together, CRISPR-Cas system makes phage gene editing simple and efficient, which can also be used to identify unknown functional genes, and expand our understanding of phage–host interactions.
The technology of artificial design and synthesis of multiple or large DNA fragments has been constantly progressed, such as Gibson assembly and Transformation-Associated Recombination (TAR).38–41 Gibson assembly is developed by Dr. Daniel Gibson et al, which allows for the successful assembly of multiple DNA fragments and the flexible and suitable construction of large DNA in vitro.42 In 2019, Nugen lab inserted a NanoLuc luciferase expression cassette into the T7 phage using Gibson assembly in vitro.43 TAR is a frequently used phage genome modification technology, which allows recombination of multiple large DNA segments in yeast artificial chromosomes (YAC) containing yeast selectable marker (HIS) and yeast centromeric locus (CEN/ARS) in yeast.44 Based on the assembly and capture of synthetic genomes into YAC, researchers have artificially modified and rebooted various Gram-positive and Gram-negative bacterial phages such as E. coli phage T3 and T7, Klebsiella phage K11, L. monocytogenes phage P35, and P. aeruginosa phages (Figure 1b).45–47 These strategies provide weapons and support for the artificial design and rebuilding of phages (Table 1).
 |
Table 1 Summary of Engineering Strategies of Phage Genomes |
Chemically Modified Phages
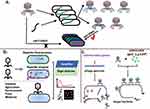
With the development of gene sequencing and synthesis technology, more and more large DNA fragments have been magnificently de novo synthesized in vitro and vivo, including phage genomes (Figure 2a). De novo genome synthesis through chemically synthesized oligonucleotides (oligos) can generate completely novel phage genome. Phix174 genome (5386 bp) was the first phage synthesized by this way in 2003.48 The synthesized oligonucleotides were gel purified, phosphorylated, annealed, and assembled in vitro, then electroporated into E. coli, followed by phage plaques checking . Even though this approach is not likely to apply to relatively large DNA molecules, it exhibits much more convenience than recombineering-based approaches. On this basis, Huiran Yeom et al presented a cell-free, low-cost, de novo gene synthesis technology called Sniper assembly for phage genome construction, and successfully obtained Acinetobacter phage AP205 genome and E. coli phage T7 genome with 89.3% and 83% success rate, respectively.49
Phages can be modified by chemical modification to improve the efficiency of sterilization or bacteria detection.50,51 Phage can be visualized through labeling its genome with fluorescence dye, such as YOYO, Cl-YO, Cl-YO-Et, Cl-YO-Bu, and the labeled phages are observed by flow cytometry and microscope.52,53 Since fluorescence dye may affect the ejection of the phage genome, dye selection is important. For example, when it was used to label phage LG1 to detect E. coli O157:H7, YOYO-1 can cross the capsid protein and bind with nucleic acids, forming the “halo-like” appearance on the cell surface rather than inside the cell.54 This is because that phage nucleic acids labeled by YOYO-1 could not be injected into bacterial cytoplasms spontaneously, unless under external force.55 SYBR-labeled phage DNA can enter the cytoplasm. In addition, dependent on nucleic acid type, available fluorescent dye is different. For example, DAPI can bind to dsDNA phage, SYBR gold binds to RNA, ssDNA, and dsDNA, and YOYO-1 binds to ssDNA and dsDNA.56
Rebooting Strategies for Engineered Phage
Phage rebooting refers to the acquisition of activated virions from the phage genome. Nowadays researchers generally rely on two ways. Firstly, phage genomic DNA is transformed into hosts or transitional hosts. Secondly, phage DNA is rebooted via cell-free transcription-translation (TXTL) systems (Table 2). The first rebooting strategy is traditional and frequently used. Phage genome is transformed into host competent cells through electro-transformation and incubated on a double-layer medium until phage plaques appear. Owing to the different cell structures between Gram-negative organisms and Gram-positive organisms, the methods are different.
 |
Table 2 Rebooting Strategies of Recombinant Phage Genomes |
In regard of Gram-negative bacterial phages, a direct chemical or electrical transformation of synthetic phage genomes into host cells requires high transformation efficiency, especially for large phage genomes. Efficient conversion protocols have been designed for certain bacteria such as E. coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae.45,46,57,58 As we know, some host genes are essential for successful phage lysis. One example is trxA in E. coli, which encodes thioredoxin, the processivity factor for T7 DNA polymerase. This gene is essential for phage replication but not essential for bacterial growth. The efficiency of plating (EOP) of T7 phage on the E. coli ∆trxA mutant was 1010-fold lower than the wild-type strain. Therefore, trxA gene could be used as a screening maker, just like an antimicrobial marker gene used in the gene knockout experiment. After recombination in vitro, if E. coli ∆trxA mutant is chosen as rebooting host, only the recombinant phage containing trxA could form plaque. This is much easier to get recombinant phage.23 Moreover, in order to produce more phage particles, the bacteria with specific restriction endonuclease gene deleted or recombinase gene expressed, sometimes could be used as a surrogate host for phage rebooting.27 This reveals that we could engineer bacterial genome to prepare the phage rebooting host when necessary. In terms of Gram-positive bacterial phages, previously universal rebooting host organisms were less, mainly restrained by DNA transformation efficiency. Recently, Listeria monocytogenes and Staphylococcus aureus cross-genus reactivation platform was developed. Kilcher et al rebooted Bacillus cereus phage TP21-L, Bacillus thuringiensis phage Bastille and Staphylococcus aureus phage 2638A and phage K besides Listeria phages P70, A511, and B035 in Listeria L-form cells.47,59 Nacyra Assad-Garcia et al rebooted Siphophage SA75, Myophage K, E. faecalis phages vB_EfaS_Ef5.1, vB_EfaS_Ef5.2, vB_EfaS_Ef5.3, vB_EfaS_Ef5.4, and vB_EfaS_Ef6.4.60
On the other hand, Cell-free TXTL system is a major improvement for phage rebooting in vitro (Figure 2b). This technique has become a suitable platform, which consists of a cell lysate or purified transcription/translation machinery and a buffer/energy mix optimized to express genes from template DNA.61–63 At first, researchers use cell extract preparation to reboot phage genome in vitro. For example, the B. subtilis phage Φ29 and T7 genome have completed the assembly in vitro utilizing the extract preparation of B. subtilis SpoA12 and E. coli extract preparation, respectively.64,65 Phage T4 was also rebooted in this cell-free reaction system.66 In recent years, Noireaux lab has successfully developed three versions of cell-free toolboxes that can reboot phage genome in vitro. At first, they completed the synthesis and rebooting of phage T7 (dsDNA, 40 kb) and ΦX174 (ssDNA, 5.4 kb) in the first version of cell-free reactions (CFRs), composing of one-third crude extract and two-thirds of water, genomes and buffer.67,68 Secondly, Version 2.0 included E. coli MazF interferase or ClpXP AAA+ proteases at the basis of Version 1.0, which can respectively degrade mRNA and high protein in order that the version has a much wider range of rates compared to Version 1.0. Rebooting of MS phage (ssRNA) was completed in this system.69 On the basis of version 2.0, two major changes of version 3.0 were made: (1) the E. coli cells were grown at 40°C instead of 37°C, (2) 60 mM maltodextrin and 30 mM d-ribose were carbohydrate source rather than only maltodextrin in cell-free reactions.70 At last, Vogele applied the cell-free sDNA (small DNA) technique (CF-sDNA) to express the native T7 phage genome in the context of cell-free protein expression by inhibiting the production of the major capsid protein of phages.71
Successful genome rebooting depends on the reaction incubation time and the potential yield of the system. Phage T4 was rebooted in cell-free reaction by optimizing biochemical settings, like concentrations of genome, Mg2+, K+ and PEG8000. Meanwhile, DNase I, EDTA, pyrophosphatase, and ATP analogs adenosine 5’-[α, β-methylene] triphosphate and adenosine 5’-[β, γ -methylene] triphosphate is crucial for bacteriophage synthesis in vitro.64,72 As a whole, the development of TXTL system not only promotes phage synthesis in vitro but also offers unique possibilities to interrogate quantitatively the links between phage gene expression, self-assembly and metabolism in the future study.
Applications of Engineered Phages
Expanding Phage Host Range
In the process of phage infection, it is necessary to recognize and bind with specific receptors on cell surfaces, which vary from bacteria to bacteria.73,74 Most of natural phages have narrow lytic host range, limiting the practical application in phage therapy.10 Therefore, it is important to broaden or alter the host range of phages by changing their genetic modules (Figure 3a). Synthetic biology techniques enable modification of phages to target different hosts by engineering and mutating phage receptor-binding proteins (RBPs), which generally are tail fibers (TFs) or tail spike proteins (TSPs).13,75,76
In 2005, Mahichi et al exchanged gp37 and gp38 at the tip of the long tail fiber of the T2 phage with the counterpart of E. coli O157:H7 specific phage PP01. The antibacterial spectrum of recombinant phage T2ppD1 was the same as that of PP01, instead of T2, and the adsorption rate was weaker than PP01.77,78 In 2015, E. coli phage T7, whose major host determinant is the tail fiber (gp17), was engineered by modular swapping of its tail component, targeting different hosts including E. coli, Yersinia and Klebsiella bacteria.46 The strategy is easier to achieve in well-studied phages. Even though the T7 phage cannot propagate in some hosts, it can package and transduce plasmids to those hosts. Yosef and G. Goren established a platform that extends the host range of T7 phage for DNA transduction by displaying various phage tail/tail fiber proteins. Specifically, they first respectively transformed 15 plasmids that encode different tail fiber genes that carry homologous arm with T7 phage tail fiber gene to E. coli hosts to produce various host E. coli. Then, when T7 phage infected those host E. coli, a plasmid expressing tail fiber gene could change the original tail fiber gene to assemble new phages that target their host to DNA transduction.79 This approach significantly extends the host range of phages and paves the way for advanced genetic manipulations and analyses, and provides a reference for other phage modifications. In 2019, Kevin Yehl et al developed a powerful high-throughput strategy to mutate T3 phage tail fiber protein that was identified as host-range-determining regions (HRDRs) through site-directed mutagenesis. This strategy created a huge diverse “phagebody” library and could suppress bacterial resistance.80
Besides, the phage genome containing the diaminopurine (Z) base is called Z-genome that can evade most restriction enzyme attacks of hosts. Zhou et al have reported a multienzyme system of Z-genome synthesis in Acinetobacter phage SH-Ab 15,497.81 Hence, it may expand the host spectrum when the Z-genome biosynthetic enzymes incorporate into the engineered phage genome. In brief, the engineered phages with expanded host range are able to effectively suppress bacterial resistance and may be very useful to boost phage applications.80,82
Detecting Bacterial Pathogen
Combining with phage specificity and reporter genes such as green fluorescent protein (gfp), luciferase-expressing gene (lux and luc), and bacterial ice nucleation (inaW), phages can be more easily, faster, and more reliably employed to detect organisms and substances with practical applications in medicine, food industry and environmental science (Figure 3b).83–85 For example, E. coli phage PP01 was labeled by fusing GFP with the small outer capsid proteins (SOC) and used for detection of E. coli O157:H7 in the sewage water. Adsorption of the GFP-labeled PP01 phages to the host cell surface made cells visualized under a fluorescence microscope.86 Due to the background natural fluorescence of biological samples, bioluminescence, a process that produces light through the enzymatic oxidation of chemical substrates is chosen as an optional substitute. Pulkkinen et al inserted a reporter luciferase enzyme Nano Luc (Nluc) into the T7 phage and used it to detect the presence of E. coli.43 A biosensor platform based on T4 phage encoding luminescent reporter enzymes allowed for detection of <10 cfu/100 mL of viable E. coli within 7 h.87 Additionally, with the advancement of molecular biology techniques, phages can be easily used to display foreign peptide sequences or materials on their coat proteins. Huan Peng et al modified the capsid of M13KE phage to display the receptor-binding proteins from different phages that naturally target the desired bacteria including E. coli, P. aeruginosa, V. cholerae, and Xanthomonas campestris.88 On the other hand, as the result of phage lysis, the released bacterial intracellular components, such as adenosine triphosphate (ATP), adenylate kinase (AK), and β-glucosidase, have also been used as cell markers for detection purposes.89–91 Hussain et al summarized the advantages and limitations of whole phage-based bacterial detection.92
The phage-based electrochemical biosensor is reported to detect various bacteria, such as Y. pseudotuberculosis, B. cereus and Mycobacterium smegmatis, and E. coli using metal ions like Hg (II), Na (I), Mn (II), Ca (II), Pb (II), and Zn (II).87,93,94 For example, engineered phage M13 displaying five RBPs from other filamentous phages aggregates AuNPs to detect E. coli, P. aeruginosa, V. cholerae, and two strains of the plant pathogen X. campestris.95 AuNPs act as a signal amplifier and detection of bacteria is achieved by testing shift in surface plasmon resonance (SPR) absorbance.96
Totally, phage-based biosensor opens the door to develop novel sensing devices such as detection of viruses and disease biomarkers or selective labeling systems for in vivo imaging, as well as identification of food pathogens. At present, the major problems of biosensors are sensitivity and repeatability. Although there are no commercially available phage-based sensing devices, multiple technologies have been patented.97,98
Enhancing Bactericidal Efficiency
To augment bactericidal activity of phage and combat antimicrobial resistance, various engineered phages are developed and applied (Figure 3c). Since phage-resistant bacteria often escape phage killing, some bacterium-killing genes originated from different organisms are integrated to phage genomes, which is the so-called “double insurance” strategy. James Cass et al have developed a SASPject platform which integrated small acid-soluble spore proteins (SASPs) encoding gene of Bacillus to phage genomes.99 SASP gene is ordinarily expressed only during sporulation, when SASPs are used to coat and protect the spore DNA. However, in vegetative cells, the binding of SASPs with DNA prevents replication and transcription and causes cell death.100,101 In vitro studies showed that the product SASPject PT1.2 killed 225 diverse isolates of Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA).102 The engineered phage Y2::dpoL1-C was constructed by introducing the depolymerase gene (dpoL1-C) of Erwinia amylovora phage L1 into the genome of E. amylovora phage Y2, which significantly increased the bactericidal efficiency compared to phage Y2.103
As we know, biofilm formation is “a devastating complication”, which can shield bacteria themselves from the host immune system and antimicrobial therapy and cause treatment inefficiency. The extraneous enzymes are a promising supplement to the lytic phages to withstand those complex, matrix-reinforced biofilms.8,104,105 For example, DspB, an enzyme that is produced by Actinobacillus actinomycetemcomitans, hydrolyzes β-1,6-N-acetyl-D-glucosamine, which is a crucial adhesin for biofilm formation and integrity in Staphylococcus and E. coli.106 Timothy K. Lu cloned the dspB gene into T7 genome under the control of the strong T7Φ10 promoter. The removal rate of E. coli biofilms by the engineered enzymatic phage was 99.997% that was about two orders of magnitude better than the original phage.107 Additionally, quorum-sensing (QS) is a process of bacterial cell-to-cell chemical communication, which is essential for virulence production and biofilm formation.108 A lactonase enzyme encoding gene was inserted into T7 phage genome, which endows the engineered phage capability of inhibiting biofilm formation of both P. aeruginosa and E. coli since this enzyme could quench QS system.50
In addition, synthetic M13 phagemid could be used as a delivery vector to upload genes for antimicrobial peptides (AMPs), or toxin proteins.109 Recombinant phagemid DNA is first transformed into a production strain harboring a helper plasmid to amplify abundant functional phagemids.110 Therefore, along with M13 infection against target bacteria, antimicrobial peptides (AMPs) or toxin proteins are expressed and inhibit intracellular processes, causing the death of nonlytic bacteria. For example, the platform expressing cecropin PR-39 or apidaecin Ia showed strong antagonistic ability against E. coli and could induce bacterial cell death effectively.111 The toxin networks-based expression of two copies of cecropin PR-39, apidaecin Ia, and topoisomerase inhibitor ccdB genes resulted in the robust killing of target E. coli.112 Mice treated by this recombinant phagemid had an average survival rate of 80% over the course of the experiment, compared to a survival rate of 27% in the untreated group.112,113
Besides phage genome engineering, phage proteins could also be modified to enhance bacterial killing efficiency. Ran et al developed a multi-functional antibacterial system (APNB) for the treatment of multi-drug resistant Acinetobacter baumannii and its biofilm, through coupling a Nile blue photosensitizer (NB) to the capsid protein of A. baumannii phage.114 The phage provides specificity and the photosensitizer produces reactive oxygen species (ROS). It is worthy to mention an interesting class of fluorescent dyes, AIEgens, which refer to fluorogens with aggregation-induced emission. Since the concept of AIE was proposed in 2001 by Tang and teammates,115 many AIEgens and AIEgens-based photodynamic therapy (PDT) approaches have been developed, such as tetraphenylethene (TPE), tetraphenylpyrazine (TPP),116 quinoline-malononitrile (QM),117 and a bacterial-based AIE molecule (TBP-2) delivery system.118 Xuewen He et al developed a novel chemical modification strategy for phage, integrating AIEgens with bacteriophage to form a new class of antimicrobial bioconjugates (TVP-PAP).119 When TVP-PAP is mixed with bacteria, TVP-PAP can be used for specific bacterial recognition and real-time fluorescent tracking in the absence of UV radiation, and host killing via TVP−PAP-guided ROS generation when in the presence of UV–vis spectra.119
In total, phages inspire the development of various antimicrobial materials, which are the promising supplement to conventional treatment strategies or industrial applications.8,104,105
Altering Microbiota Composition
As microbiome modulators, phages can improve microbiome structure and suppress pathogens. Although it is infrequent in a clinic, researchers confirmed its authenticity in mice models and ecological microbiome.120–124 Hsu et al constructed a mouse model carrying human gut commensal bacteria and tested the influence of lytic E. coli phage T4, C. sporogenes phage F1, phage B. fragilis B40-8, and E. faecalis phage VD13 on to the gut microbiome. The results showed that phages not only directly impacted targeted bacteria but also resulted in cascading effects on other bacterial species.124 Zheng reported that Fusobacterium nucleatum lytic phage-guided nanoparticles reduced the side effects of chemotherapy drugs and promoted the proliferation of endogenic Clostridium butyricum in mouse models of colorectal cancer.125 The strategy provides a new way for the development and application in cancer treatment. In addition, phages have also been utilized as a precise regulatory tool to control the natural rhizosphere microbiomes. Phages were used to decrease the incidence of Ralstonia solanacearum by up to 80% in tomatoes, which did not affect the existing rhizosphere microbiota.126
Conclusions and Perspectives
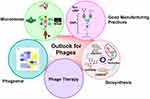
With the quick spread of AMR and the slow pace of new antibiotics discovery, the design and development of phage or phage functional elements, as novel and alternative antibacterial therapies, detection and diagnostic tools, have been proposed and implemented (Figure 4).127–129 As well, developing a more convenient phage display technique is needed for protein engineering. In this new journey, the road ahead is still long. Considering the limitations of applications such as their narrow host range, immune toxicity, and low infection efficiency, it is time-consuming and laborious to select suitable phages from the natural phage library. Under “design-build-test-learn” theory, synthetic biologists manipulate phage genomes bottom-up by means of evolving molecular biology methods, such as recombination tools, large fragment cloning technology, CRISPR and base editing, cheap synthetic genes, and cell-free systems, to obtain engineered phages with specific functions to support pathogen defense, drug delivery, bacterial detection, and material science.
Engineering of phage is encouraging. The huge diversity of phage types and gene pools in nature is our valuable gift. So far, phage synthetic biology has only utilized a small part of the existing phage types, and the potential of the vast arsenal is far from being tapped. For commercial production and application of the engineered phages in “outside-The-lab” spaces in the future, challenges still remain, such as long-term stability, storage and production conditions, and production cost.130 With the innovation of technology and the combination of multi-disciplines, it is believed that it will come true to develop robust, cost-effective, safe, and efficacious platforms that can translate more and more suitable engineered phages in the lab into real-world applications.
Funding
This study was supported by grants from Science & Technology Fundamental Resources Investigation Program (2022FY101100) and the National Natural Science Foundation of China (32170114 and 82272340).
Disclosure
The authors declare no conflicts of interest.
References
1. Merril CR, Scholl D, Adhya SL. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov. 2003;2(6):489–497.
2. Salmond GP, Fineran PC. A century of the phage: past, present and future. Nat Rev Microbiol. 2015;13(12):777–786.
3. Correa AMS, Howard-Varona C, Coy SR, Buchan A, Sullivan MB, Weitz JS. Revisiting the rules of life for viruses of microorganisms. Nat Rev Microbiol. 2021;19:501–513.
4. Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK. Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev. 2016;80(3):523–543.
5. Kilcher S, Loessner MJ. Engineering bacteriophages as versatile biologics. Trends Microbiol. 2019;27(4):355–367.
6. Peng H, Chen IA. Phage engineering and the evolutionary arms race. Curr Opin Biotechnol. 2021;68:23–29.
7. Khan Mirzaei M, Deng L. New technologies for developing phage-based tools to manipulate the human microbiome. Trends Microbiol. 2021;30(2):131–142.
8. Hansen MF, Svenningsen SL, Røder HL, Middelboe M, Burmølle M. Big impact of the tiny: bacteriophage-bacteria interactions in biofilms. Trends Microbiol. 2019;27(9):739–752.
9. Citorik RJ, Mimee M, Lu TK. Bacteriophage-based synthetic biology for the study of infectious diseases. Curr Opin Microbiol. 2014;19:59–69.
10. Brown R, Lengeling A, Wang B. Phage engineering: how advances in molecular biology and synthetic biology are being utilized to enhance the therapeutic potential of bacteriophages. Quant Biol. 2017;5(1):42–54.
11. Meng F, Ellis T. The second decade of synthetic biology: 2010–2020. Nat Commun. 2020;11(1):5174.
12. Voigt CA. Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat Commun. 2020;11(1):6379.
13. Dams D, Brøndsted L, Drulis-Kawa Z, Briers Y. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem Soc Trans. 2019;47(1):449–460.
14. Lu TK, Bowers J, Koeris MS. Advancing bacteriophage-based microbial diagnostics with synthetic biology. Trends Biotechnol. 2013;31(6):325–327.
15. Pizarro-Bauerle J, Ando H. Engineered bacteriophages for practical applications. Biol Pharm Bull. 2020;43(2):240–249.
16. Deng X, Wang L, You X, Dai P, Zeng Y. Advances in the T7 phage display system (Review). Mol Med Rep. 2018;17(1):714–720.
17. Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH. Basics of antibody phage display technology. Toxins. 2018;10(6):236.
18. Lu TK, Koeris MS. The next generation of bacteriophage therapy. Curr Opin Microbiol. 2011;14(5):524–531.
19. Lobocka M, Dabrowska K, Gorski A. Engineered bacteriophage therapeutics: rationale, challenges and future. BioDrugs. 2021;35(3):255–280.
20. Marinelli LJ, Hatfull GF, Piuri M. Recombineering: a powerful tool for modification of bacteriophage genomes. Bacteriophage. 2012;2(1):5–14.
21. Pearson RE, Jurgensen S, Sarkis GJ, et al. Construction of D29 shuttle phasmids and luciferase reporter phages for detection of mycobacteria. Gene. 1996;183(1–2):129–136.
22. Le S, He X, Tan Y, et al. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS One. 2013;8(7):e68562.
23. Udi Qimron BM, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. PNAS. 2006;103(50):19039–19044.
24. Tran NQ, Udi Qimron LFR. Gene 1.7 of bacteriophage T7 confers sensitivity of phage growth to dideoxythymidine. PNAS. 2008;105(27):9373–9378.
25. Grigonyte AM, Paul CH, MacDonald R, et al. Comparison of CRISPR and marker-based methods for the engineering of phage T7. Viruses. 2020;12(2):193.
26. Feher T, Karcagi I, Blattner FR, Pósfai G. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb Biotechnol. 2012;5(4):466–476.
27. Jensen JD, Parks AR, Adhya S, Rattray AJ, Court DL. lambda recombineering used to engineer the genome of phage T7. Antibiotics. 2020;9(11):805.
28. Marinelli LJ, Piuri M, Swigoňová Z, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One. 2008;3(12):e3957.
29. Duong MM, Carmody CM, Ma Q, Peters JE, Nugen SR. Optimization of T4 phage engineering via CRISPR/Cas9. Sci Rep. 2020;10(1):18229.
30. Schilling T, Dietrich S, Hoppert M, Hertel R. A CRISPR-Cas9-based toolkit for fast and precise in vivo genetic engineering of bacillus subtilis phages. Viruses. 2018;10(5):241.
31. Shen J, Zhou J, Chen GQ, Xiu ZL. Efficient genome engineering of a virulent Klebsiella bacteriophage using CRISPR-Cas9. J Virol. 2018;92(17):e00534–18.
32. Lemay ML, Tremblay DM, Moineau S. Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth Biol. 2017;6(7):1351–1358.
33. Martel B, Moineau S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014;42(14):9504–9513.
34. Bari SMN, Walker FC, Cater K, Aslan B, Hatoum-Aslan A. Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synth Biol. 2017;6(12):2316–2325.
35. Dong J, Chen C, Liu Y, Zhu J, Li M, Rao VB, Tao P. Engineering T4 bacteriophage for in vivo display by type V CRISPR-cas genome editing. ACS Synth Biol. 2021;10(10):2639–2648.
36. Kiro R, Shitrit D, Qimron U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014;11(1):42–44.
37. Box AM, McGuffie MJ, O’Hara BJ, Seed KD. Functional analysis of bacteriophage immunity through a type I-E CRISPR-cas system in vibrio cholerae and its application in bacteriophage genome engineering. J Bacteriol. 2016;198(3):578–590.
38. Anderson J, Dueber JE, Leguia M, et al. BglBricks: a flexible standard for biological part assembly. J Biol Eng. 2010;4(1):1–12.
39. Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4(5):e5553.
40. Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, et al. GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013;162(3):1618–1631.
41. de Kok S, Stanton LH, Slaby T, et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol. 2014;3(2):97–106.
42. Gibson DG, Young L, Chuang RY, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345.
43. Pulkkinen EM, Hinkley TC, Nugen SR. Utilizing in vitro DNA assembly to engineer a synthetic T7 Nanoluc reporter phage for Escherichia coli detection. Integrat Biol. 2019;11(3):63–68.
44. Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3(3):371–377.
45. Pires DP, Monteiro R, Mil-Homens D, Fialho A, Lu TK, Azeredo J. Designing P. aeruginosa synthetic phages with reduced genomes. Sci Rep. 2021;11(1):2164.
46. Ando H, Lemire S, Pires DP, Lu TK. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015;1(3):187–196.
47. Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc Natl Acad Sci USA. 2018;115(3):567–572.
48. Smith HO, Cynthia Pfannkoch CAHI, Hutchison CA, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: Ø X174 bacteriophage from synthetic oligonucleotides. PNAS. 2003;100(26):15440–15445.
49. Yeom H, Ryu T, Lee AC, Cell-free bacteriophage genome synthesis using low-cost sequence-verified array-synthesized oligonucleotides. ACS Synth Biol. 2020;9(6):1376–1384.
50. Anam GB, Yadav S, Ayyaru S, Ahn YH. Nanocomposite membrane integrated phage enrichment process for the enhancement of high rate phage infection and productivity. Biochem Eng J. 2020;163:107740.
51. Malik DJ. Approaches for manufacture, formulation, targeted delivery and controlled release of phage-based therapeutics. Curr Opin Biotechnol. 2021;68:262–271.
52. Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53(1):9–18.
53. Vus K, Tarabara U, Balklava Z, et al. Association of novel monomethine cyanine dyes with bacteriophage MS2: a fluorescence study. J Mol Liq. 2020;302:112569.
54. Goodridge L, Mansel Griffiths JC. Development and characterization of a fluorescent-bacteriophage assay for detection of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65(4):1397–1404.
55. Mattias Karlsson KN, Davidson MJ, Nolkrantz K, Davidson MJ, et al. Electroinjection of colloid particles and biopolymers into single unilamellar liposomes and cells for bioanalytical applications. Anal Chem. 2000;72(23):5857–5862.
56. Mosier-Boss PA, Lieberman SH, Andrews JM, Rohwer FL, Wegley LE, Breitbart M. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl Spectrosc. 2003;57(9):1138–1144.
57. Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. A fully decompressed synthetic bacteriophage oX174 genome assembled and archived in yeast. Virology. 2012;434(2):278–284.
58. Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1(p. 2005):0018.
59. Studer P, Staubli T, Wieser N, et al. Proliferation of Listeria monocytogenes L-form cells by formation of internal and external vesicles. Nat Commun. 2016;7:13631.
60. Nacyra Assad-Garcia RDS, Rachel Buzzeo B. Cross-genus “boot-up” of synthetic bacteriophage in staphylococcus aureus by using a new and efficient DNA Transformation method. Appl Environ Microbiol. 2022;88(3):e01486–21.
61. Garenne D, Noireaux V. Cell-free transcription-translation: engineering biology from the nanometer to the millimeter scale. Curr Opin Biotechnol. 2019;58:19–27.
62. Silverman AD, Karim AS, Jewett MC. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21(3):151–170.
63. Garenne D, Bowden S, Noireaux V. Cell-free expression and synthesis of viruses and bacteriophages: applications to medicine and nanotechnology. Curr Opin Syst Biol. 2021;28:100373.
64. Bjornsti MA, Reilly BE, Anderson DL. In vitro assembly of the Bacillus subtilis bacteriophage Ø 29. Proc Nad Acad Sci. 1981;78(9):5861–5865.
65. Bjornsti MA, Reilly BE, Anderson DL. Morphogenesis of bacteriophage phi29 of bacillus subtilis: DNA-gp3 intermediate in in vivo and in vitro assembly. J Virol. 1982;41(2):508–517.
66. Rustad M, Eastlund A, Jardine P, Noireaux V. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth Biol. 2018;3(1):ysy002.
67. Shin J, Jardine P, Noireaux V. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth Biol. 2012;1(9):408–413.
68. Rustad M, Eastlund A, Marshall R, Jardine P, Noireaux V. Synthesis of infectious bacteriophages in an E. coli-based cell-free expression system. J Vis Exp. 2017;2017(126):56144.
69. Garamella J, Marshall R, Rustad M, Noireaux V. The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth Biol. 2016;5(4):344–355.
70. Garenne D, Thompson S, Brisson A, Khakimzhan A, Noireaux V. The all-E. coliTXTL toolbox 3.0: new capabilities of a cell-free synthetic biology platform. Synth Biol. 2021;6(1):ysab017.
71. Vogele K, Falgenhauer E, von Schonberg S, Simmel FC, Pirzer T. Small antisense DNA-based gene silencing enables cell-free bacteriophage manipulation and genome replication. ACS Synth Biol. 2021;10(3):459–465.
72. Masker WE, Kuemmerle NB, Allison DP. In vitro packaging of bacteriophage T7 DNA synthesized in vitro. J Virol. 1978;27(1):149–163.
73. Dennehy JJ, Abedon ST. Adsorption: phage acquisition of bacteria. Bacteriophages. 2020;2020:1–25.
74. Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. 2016;363:4.
75. Lenneman BR, Fernbach J, Loessner MJ, Lu TK, Kilcher S. Enhancing phage therapy through synthetic biology and genome engineering. Curr Opin Biotechnol. 2020;68:151–159.
76. Taslem Mourosi J, Awe A, Guo W, et al. Understanding bacteriophage tail fiber interaction with host surface receptor: the key “blueprint” for reprogramming phage host range. Int J Mol Sci. 2022;23:20.
77. Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol. 2005;115(1):101–107.
78. Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett. 2009;295(2):211–217.
79. Yosef I, Goren MG, Globus R, Molshanski-Mor S, Qimron U. Extending the host range of bacteriophage particles for DNA transduction. Mol Cell. 2017;66(5):721–728 e3.
80. Yehl K, Lemire S, Yang AC, et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell. 2019;179(2):459–469.
81. Zhou Y, Xu X, Wei Y, et al. A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science. 2021;372(6541):512–516.
82. Russell J, Bikard D. Learning from antibodies: phage host-range engineering. Cell Host Microbe. 2019;26(4):445–446.
83. Machera SJ, Niedziółka-Jönsson J, Szot-Karpińska K. Phage-based sensors in medicine: a review. Chemosensors. 2020;8(3):61.
84. Bayat F, Didar TF, Hosseinidoust Z. Emerging investigator series: bacteriophages as nano engineering tools for quality monitoring and pathogen detection in water and wastewater. Environ Sci. 2021;8(2):367–389.
85. Schofield DA, Sharp NJ, Westwater C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage. 2012;2(2):105–283.
86. Oda M, Morita M, Unno H, Tanji Y. Rapid detection of Escherichia coli O157: h7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl Environ Microbiol. 2004;70(1):527–534.
87. Zurier HS, Duong MM, Goddard JM, Nugen SR. Engineering biorthogonal phage-based nanobots for ultrasensitive, in situ bacteria detection. ACS Appl Bio Mater. 2020;3(9):5824–5831.
88. Peng H, Chen IA. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano. 2019;13(2):1244–1252.
89. Yemini M, Levi Y, Yagil E, Rishpon J. Specific electrochemical phage sensing for Bacillus cereus and Mycobacterium smegmatis. Bioelectrochemistry. 2007;70(1):180–184.
90. Blasco R, Murphy MJ, Sanders MF, Squirrell DJ. Specific assays for bacteria using phage mediated release of adenylate kinase. J Appl Microbiol. 2010;84(4):661–666.
91. Gupta V, Saxena HM. A new bacteriophage based luminescence assay for diagnosis of brucellosis. Indian J Exp Biol. 2017;55(5):296–302.
92. Hussain W, Ullah MW, Farooq U, Aziz A, Wang S. Bacteriophage-based advanced bacterial detection: concept, mechanisms, and applications. Biosens Bioelectron. 2021;177:112973.
93. Yang Q, Deng S, Xu J, et al. Poly (indole-5-carboxylic acid)/reduced graphene oxide/gold nanoparticles/phage-based electrochemical biosensor for highly specific detection of Yersinia pseudotuberculosis. Mikrochim Acta. 2021;188(4):107.
94. Manivannan S, Park S, Jeong J, Kim K. Aggregation-free optical and colorimetric detection of Hg (II) with M13 bacteriophage-templated Au nanowires. Biosens Bioelectron. 2020;161:112237.
95. Peng H, Borg RE, Nguyen AB, Chen IA. Chimeric phage nanoparticles for rapid characterization of bacterial pathogens: detection in complex biological samples and determination of antibiotic sensitivity. ACS Sens. 2020;5(5):1491–1499.
96. Kim T, Hyeon T. Applications of inorganic nanoparticles as therapeutic agents. Nanotechnology. 2014;25(1):012001.
97. Ramasamy RP, Zhou Y. Bacteriophage-based electrochemical bacterial sensors, systems, and methods; 2018.
98. Jeff A, Conrad AJ. Biologic machines for the detection of biomolecules; 2015.
99. Barnard AML, Cass JA. Targetable nano-delivery vehicles to deliver anti-bacterial small acid-soluble spore protein (SASP) genes. Emerg Top Life Sci. 2021;5(5):637–641.
100. Setlow P. Spore resistance properties. Microbiology Spectrum. 2014;2(5):TBS-0003–2012.
101. Mohr SC, Sokolov NV, Chaomei HE. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Natl Acad Sci USA. 1991;88:77–81.
102. Cass J, Barnard A, Fairhead H. Engineered bacteriophage as a delivery vehicle for antibacterial protein, SASP. Pharmaceuticals. 2021;14(10):1038.
103. Born Y, Fieseler L, Thöny V, Leimer N, Duffy B, Loessner MJ. Engineering of bacteriophages Y2:: dpoL1-Cand Y2:: luxABfor efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl Environ Microbiol. 2017;83:12.
104. Chan BK, Abedon ST. Bacteriophages and their enzymes in biofilm control. Curr Pharm Des. 2015;21(1):85–99.
105. Sasikala D, Srinivasan P. Characterization of potential lytic bacteriophage against Vibrio alginolyticus and its therapeutic implications on biofilm dispersal. Microb Pathog. 2016;101:24–35.
106. Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75(1):125–132.
107. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104(27):11197–11202.
108. Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11):a012427.
109. Russel M. Genetic analysis of the filamentous bacteriophage packaging signal and of the proteins that interact with it. Am Soc Microbiol. 1989;63(8):3284–3295.
110. Chasteen L, Ayriss J, Pavlik P, Bradbury AR. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34(21):e145.
111. Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250.
112. Krom RJ, Bhargava P, Lobritz MA, Collins JJ. Engineered phagemids for nonlytic, targeted antibacterial therapies. Nano Lett. 2015;15(7):4808–4813.
113. Wu WH, Zhang MP, Zhang F, et al. The role of programmed cell death in streptozotocin-induced early diabetic nephropathy. Tumor Biol. 1996;99:129–141.
114. Ran B, Yuan Y, Xia W, et al. A photo-sensitizable phage for multidrug-resistant Acinetobacter baumannii therapy and biofilm ablation. Chem Sci. 2021;12(3):1054–1061.
115. Luo J, Xie Z, Lam JW, et al. Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole. Chem Commun. 2001;2001(18):1740–1741.
116. Chen M, ZhongáTang B. Tetraphenylpyrazine-based AIEgens: facile preparation and tunable light emission. Chem Sci. 2015;6(3):1932–1937.
117. Li Y, Shao A, Wang Y, et al. Morphology-tailoring of a red AIEgen from microsized rods to nanospheres for tumor-targeted bioimaging. Adv Mater. 2016;28(16):3187–3193.
118. Zhu D, Zhang J, Luo G, Duo Y, Tang BZ. Bright bacterium for hypoxia-tolerant photodynamic therapy against orthotopic colon tumors by an interventional method. Adv Sci. 2021;8(15):e2004769.
119. He X, Yang Y, Guo Y, et al. Phage-guided targeting, discriminative imaging, and synergistic killing of bacteria by AIE bioconjugates. J Am Chem Soc. 2020;142(8):3959–3969.
120. Kahrstrom CT. Microbiome: with a little help from my phage friends. Nat Rev Microbiol. 2013;11(8):507.
121. Dahlman S, Avellaneda-Franco L, Barr JJ. Phages to shape the gut microbiota? Curr Opin Biotechnol. 2021;68:89–95.
122. Khan Mirzaei M, Deng L. Sustainable Microbiome: a symphony orchestrated by synthetic phages. Microb Biotechnol. 2021;14(1):45–50.
123. Zhang Y, Li CX, Zhang XZ. Bacteriophage-mediated modulation of microbiota for diseases treatment. Adv Drug Deliv Rev. 2021;176:113856.
124. Hsu BB, Gibson TE, Yeliseyev V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe. 2019;25(6):803–814 e5.
125. Zheng DW, Dong X, Pan P, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng. 2019;3(9):717–728.
126. Wang X, Wei Z, Yang K, et al. Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol. 2019;37(12):1513–1520.
127. Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232.
128. Torres-Barcelo C, Hochberg ME. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016;24(4):249–256.
129. Zhang W, Wu Q. Applications of phage-derived RNA-based technologies in synthetic biology. Synth Syst Biotechnol. 2020;5(4):343–360.
130. Brooks SM, Alper HS. Applications, challenges, and needs for employing synthetic biology beyond the lab. Nat Commun. 2021;12(1):1390.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.