Back to Journals » OncoTargets and Therapy » Volume 12
Aberrant Expression of miR-592 Is Associated with Prognosis and Progression of Renal Cell Carcinoma
Authors Lv X, Shen J, Guo Z, Kong L, Zhou G, Ning H
Received 19 August 2019
Accepted for publication 5 November 2019
Published 18 December 2019 Volume 2019:12 Pages 11231—11239
DOI https://doi.org/10.2147/OTT.S227834
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Takuya Aoki
Xianbao Lv,1,* Jingang Shen,1,* Zhen Guo,2,* Lingwei Kong,1 Guangchun Zhou,1 Hao Ning3
1Department of Urology, Chengwu People’s Hospital, Heze, Shandong, 274200, People’s Republic of China; 2Department of Urology, Central Hospital Affiliated Shandong First Medical University, Jinan, Shandong, 250033, People’s Republic of China; 3Department of Urology, Shandong Provincial Hospital, Jinan, Shandong, 250021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hao Ning
Department of Urology Surgery, Shandong Provincial Hospital, No. 324, Jingwu Road, Jinan, Shandong 250021, People’s Republic of China
Tel +86 531 68776310
Email [email protected]
Purpose: MicroRNAs have recently reported playing a vital role in the development of cancers. However, the role of miR-592 in renal cell carcinoma (RCC) has not been explored. In this study, the potential role of miR-592 was investigated in RCC.
Patients and methods: The expression of miR-592 was evaluated in RCC tissues and cell lines using qRT-PCR assays. The Kaplan-Meier analysis and Cox proportional hazards model analysis was used to analyze the prognostic value of miR-592 in RCC. The effects of miR-592 on cell proliferation, migration, and invasion were determined by cell counting kit-8 (CCK-8) and Transwell assays in vitro.
Results: The results showed that miR-592 was significantly increased both in RCC tissues and cell lines. Overexpression of miR-592 was significantly associated with lymph node metastasis, TNM stage, and poor overall survival. And functional studies in two RCC cell lines (786-O and Caki-1) have shown that overexpression of miR-592 promoted cell proliferation, migration, and invasion, while silence of miR-592 inhibited cell proliferation, migration, and invasion. SPRY2 was a direct target of miR-592.
Conclusion: Overall, overexpression of miR-592 may be a prognostic biomarker and therapeutic strategy for patients with RCC, which is correlated with the progression of RCC.
Keywords: miR-592, renal cell carcinoma, prognosis, proliferation, migration, invasion
Introduction
Renal cell carcinoma (RCC), a common human kidney cancer, represents approximately 3% of all adult malignancies, which is the third leading cause of death in urological cancers.1,2 There are five histological types of RCC, mainly include clear cell RCC and papillary RCC.3 At present, surgery remains the mainstay of treatment for located RCC.4 However, approximately one-third of RCC patients have had metastasis at initial diagnosis and were resistant to chemotherapy and radiotherapy.5,6 Although considerable advances have been made in the therapeutic strategies, these patients at advanced stages usually have an extremely poor prognosis with a median survival of approximately 13 months.7 Therefore, it is urgently to identify novel therapeutic targets and accurate prognostic biomarkers to improve treatment and prognosis for RCC patients.
MicroRNAs (miRNAs) are a group of small noncoding RNA molecules (approximately 19–22 nucleotides in length), known to negatively regulate their target genes at the post-transcriptional level by binding to the 3ʹ-UTR of target mRNAs.8 Increasing evidence has been identified miRNAs involved in diverse cellular processes, such as cell proliferation, differentiation, invasion, and apoptosis.9–11 Recently, numerous studies demonstrated that aberrant miRNAs expression was closely related to the development and progression of cancers and functioned as promising therapeutic targets or biomarkers for cancers.12–14 miR-592 has been reported aberrantly expressed in a variety of cancers, such as glioma,15 hepatocellular carcinomas,16 and colorectal cancer.17 Although a previous literature search study showed that miR-592 was upregulated in RCC,18 its role and clinical significance have not yet been clarified in RCC.
In the present study, we explored the expression levels of miR-592 in RCC tissues and cells. The relationship between the expression of miR-592 and clinicopathological characteristics of RCC patients was also investigated. What’s more, we explored the clinical significance of miR-592 and its role in RCC and implored it is as a promising biomarker and therapeutic target for RCC.
Materials and Methods
Patients and Tissue Specimens
Ethical approval for this study was obtained from the Research Ethics Committee of Chengwu People’s Hospital. All patients signed written informed consent prior to sampling. The paired RCC tissue specimens and adjacent normal tissue specimens were obtained from 114 RCC patients who underwent surgical resection at Chengwu People’s Hospital between February 2011 and December 2013. Tissues were immediately snap-frozen in liquid nitrogen until RNA extraction. No patients received preoperative chemotherapy or radiotherapy prior to surgical resection. The corresponding clinical characteristics of RCC patients (shown in Table 1) and 5-year follow-up information were collected and recorded.
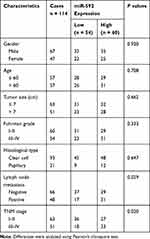 |
Table 1 Clinicopathological Characteristics of RCC Patients and Their Association with miR-592 Expression |
Cell Culture and Transient Transfection
Human renal carcinoma cell lines 786-O, ACHN, Caki-1, 769-P and normal renal epithelial cells (HK-2) were all purchased from the Institute of Cell Research, Chinese Academy of Sciences (Shanghai, China). 786-O, ACHN, Caki-1, and 769-P cells were cultured in RPMI-1640 (HyClone, Logan, UT, USA) supplemented with 10% FBS (HyClone). HK-2 cells were cultured in keratinocyte serum-free medium (K-SFM; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (HyClone). All cells were cultured in a humidified incubator with 5% CO2 at 37°C.
For overexpression and downregulation of miR-592, the miR-592 mimic, mimic negative control (mimic NC), miR-592 inhibitor, and inhibitor NC used in this study were chemically synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from RCC tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. The RNA quality and quantity were verified with a NanoDrop 1000 (Thermo Fisher Scientific). Then, the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) was used to synthesize cDNA from total RNA following the manufacturer’s instructions. Subsequently, qRT-PCR was performed to measure miR-592 expression using a TaqMan MicroRNA Assay kit (Thermo Fisher Scientific) on an ABI 7500 PCR System. The relative expression levels of miR-592 were calculated using 2−ΔΔCt methods and normalized to those of U6.
Cell Proliferation Assay
The proliferative abilities of RCC cells were measured using a Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China). In brief, approximately 3 × 103 transfected cells in culture medium were plated onto 96-well plates and incubated at 37°C with 5% CO2. At 0, 24, 48, 72 h, 10 μL CCK-8 reagent was added into each well and the absorbance values were read at 450 nm with a microplate reader.
Cell Migration and Invasion Assays
The migration and invasion abilities of RCC cells were measured using a 24-well Transwell plate (8 μm; BD Biosciences, San Jose, CA). Chambers without or with Matrigel (BD Biosciences, San Jose, CA, USA) were used to determine the migration ability and invasion ability of RCC cells, respectively. Serum-free culture medium containing 2 × 104 transfected cells was added into the upper chamber and culture medium supplemented with 10% FBS was added to the lower chamber as a chemoattractant. Following culturing at 37°C for 24 h, cells that migrated to or invaded the bottom chambers were fixed, stained, and counted in five random fields under a microscope.
Bioinformatics Analysis
Bioinformatics prediction was used to analyze the putative targets of miR-592 using publicly available algorithms, the TargetScan (http://www.targetscan.org/).
Luciferase Reporter Assay
For dual reporter luciferase assay, the SPRY2 3ʹ-UTR oligonucleotides containing the wild type (Wt) or mutant (Mut) miR-592 binding sites were constructed by GenePharma Co., Ltd (Shanghai, China). The SPRY2 Wt or Mut 3ʹ-UTR was cloned into the pGL3 luciferase reporter vector (Promega, Madison, WI, USA). Caki-1 cells were seeded in 24-well plates and co-transfected with Wt-SPRY2-3ʹ-UTR or Mut-SPRY2 and miR-592 mimic, mimic NC, miR-592 inhibitor, or inhibitor NC using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. After transfection for 24 h, transfected cells were collected and luciferase activity was measured using a dual reporter assay system (Promega, Madison, WI, USA) that was normalized to Renilla luciferase activity.
Statistical Analyses
All statistical analyses were performed using SPSS 20.0 (SPSS Inc., Chicago, IL, USA) or GraphPad 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Each experiment was performed at least three times. Data were presented as the mean ± SD. Kruskal–Wallis test was used to confirm whether the data are parametric. The statistical differences between two groups were analyzed using a 2-tailed Student’s t-test. A one-way ANOVA followed by Tukey’s post-hoc test was used for multiple groups comparison. The association between miR-592 expression and clinical characteristics of patients was analyzed using Pearson’s chi-square test. The Kaplan-Meier analysis was used to estimate the overall survival of patients with different miR-592 expression. Multivariate Cox regression analysis was adopted to analyze prognostic factors. Differences with P < 0.05 were considered to be statistically significant.
Results
Expression of miR-592 in RCC Tissues and Cell Lines
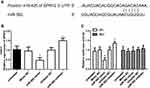
To clarify whether the miR-592 expression is associated with RCC progression, RCC tissues and adjacent normal tissues, as well as RCC cells were analyzed using qRT-PCR. Firstly, miR-592 expression was detected by qRT-PCR in 114 paired fresh tumor tissues and adjacent normal tissues. Compared with adjacent normal tissues, miR-592 expression was significantly higher in tumor tissues (P < 0.001, Figure 1A). Then we examined the miR-592 expression in RCC cells. As shown in Figure 1B, miR-592 expression was upregulated in RCC cells compared with that in normal renal epithelial HK-2 cells (P < 0.001).
Relationship Between Clinicopathological Characteristics and miR-592 Expression in RCC Patients
We classified the patients into a low miR-592 expression group and a high miR-592 expression group according to the relative mean expression level of miR-592 in RCC tissues. Next, the patients’ clinicopathological characteristics of RCC patients were analyzed using the chi-square test (Table 1). The analysis results showed that miR-592 expression was significantly associated with lymph node metastasis (P = 0.029) and TNM stage (P = 0.020). However, no significant association was found between miR-592 expression and other characteristics, such as gender, age, tumor size, and differentiation (P > 0.05).
Upregulation of miR-592 Is Associated with Poor Prognosis in RCC Patients
In order to assess the prognostic value of miR-592, Kaplan-Meier survival curves and log rank tests were performed. The results showed that the overall survival of patients with high miR-592 expression was significantly worse than those with low miR-592 expression (P = 0.009, Figure 2). Further multivariate Cox regression analyses showed that miR-592 expression (HR = 2.265, 95% CI: 1.048–4.899, P = 0.038) and TNM stage (HR = 2.134, 95% CI: 1.006–4.527, P = 0.048) were associated with overall survival and independent prognostic factors for RCC (Table 2).
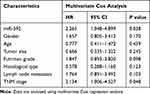 |
Table 2 Multivariate Cox Analyses of Overall Survival of RCC Patients |
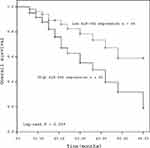 |
Figure 2 Kaplan-Meier curves for patients with RCC expressing high and low levels of miR-592 (log rank test: P = 0.009). |
Upregulation of miR-592 Promotes Proliferation, Migration, and Invasion of RCC Cells in vitro
To further evaluate the functional role of miR-592 in RCC, the impact of its overexpression or downregulation on the proliferation, migration, and invasion of RCC cells was investigated. 786-O and Caki-1 cells were selected for functional experiments because of their higher expression levels of miR-592, which were transfected with miR-592 mimics or miR-592 inhibitors. The overexpression or downregulation of miR-592 was confirmed by qRT-PCR in 786-O and Caki-1 cells following transfection with miR-592 mimics or inhibitors (P < 0.001, Figure 3A). A CCK-8 assay was used to investigate the proliferative ability of RCC cells. As shown in Figure 3B, the proliferative abilities of 786-O and Caki-1 were significantly promoted by miR-592 overexpression, while the proliferative abilities were markedly inhibited by the knockdown of miR-592, compared with that in untreated cells (P < 0.05). The Transwell assays were performed to determine the effect of miR-592 on cell migration and invasion. The results revealed that upregulation of miR-592 accelerated the migration and invasive capabilities of 786-O and Caki-1 cells, while downregulation of miR-592 resulted in attenuation of migration and invasive capabilities, compared with that of untreated cells (P < 0.01, Figure 4).
SPRY2 Is a Direct Target of miR-592 in RCC Cells
TargetScan was used to predict the potential targets of miR-592. The results showed that SPRY2 was a predicted target of miR-592 and there was a binding site of miR-592 in the 3ʹ-UTR of SPRY2 (Figure 5A). Then, the luciferase reporter assay was performed with RCC Caki-1cells, which were transfected with luciferase constructs containing Wt-3ʹ-UTR and Mut-3ʹ-UTR of SPRY2. Overexpression of miR-592 reduced the expression of SPRY2, while the miR-592 inhibitor increased the SPRY2 expression (P < 0.05, Figure 5B). Furthermore, dual-luciferase assays showed that the relative luciferase activity was significantly decreased in Caki-1 cells cotransfected with miR-592 mimic and Wt SPRY2 vector, while no significant change in luciferase activity was observed in the Mut SPRY2 group (P < 0.05, Figure 5C).
Discussion
Increasing evidence has demonstrated that miRNAs are frequently aberrantly expressed in various cancers and are associated with tumor progression and survival of various cancers.19–21 For instance, the miR-629-3p expression is upregulated in human lung adenocarcinoma tissues and cell lines, and it promotes cell proliferation and predicts poor survival in lung adenocarcinoma.22 In RCC, a number of miRNAs have identified associated with the prognosis of patients and functioned as prognostic biomarkers,23 such as miR-30a24 and miR-660-5p.25 A recent study demonstrated that the upregulation of miR-142-3p is correlated with poor prognosis of RCC patients and may be a prognostic biomarker.26 Therefore, investigation of cancer-associated miRNAs that are crucial for the progression of RCC may provide more therapeutic targets and prognosis improvement for RCC patients.
In the present study, miR-592 may exhibit tumor oncogene action in RCC. The key findings of the present study were as follows. The miR-592 expression was significantly upregulated in RCC tissues and cell lines. High miR-592 expression was markedly associated with lymph node metastasis and TNM stages. In addition, increased miR-592 expression was associated with the poor prognosis of RCC patients. Moreover, overexpression of miR-592 promoted cell proliferation, migration, and invasion of RCC cells. Overall, these results suggested that miR-592 may be a potential prognostic biomarker or therapeutic target for patients with RCC.
Previous studies have reported that miR-592 was aberrantly expressed and associated with the progression of several cancer types.15–17 For instance, miR-592 functions as a suppressive role in breast cancer and inhibits cell proliferation, clone formation, migration, and invasion in breast cancer.27 The miR-592 expression is downregulated in non-small cell lung cancer (NSCLC) and plays tumor suppressor functions on NSCLC proliferation, colony formation, migration, and invasion by targeting SOX9.28 On the contrary, in some other studies, miR-592 plays an oncogenic role in tumorigenesis of several cancers, such as gastric cancer,29 colorectal cancer,30 and prostate cancer.31 Considering the expression pattern of miR-592 in different cancers, we deemed that miR-592 may act as either an oncogene or a tumor suppressor gene depending on cancer type. In the present study, miR-592 was significantly upregulated in RCC tissues and cell lines, compared with that in adjacent normal tissues and normal HK-2 cells, respectively. Our findings are similar to previous studies in which miR-592 contributed to the oncogenesis of gastric cancer, colorectal cancer, and prostate cancer.29–31 Moreover, high expression of miR-592 was found significantly associated with lymph node metastasis and TNM stages. These results suggested miR-592 may be associated with the development of RCC. Kaplan-Meier curve analysis and multivariate Cox analysis results showed miR-592 expression was a prognostic factor for overall survival of RCC patients. The above outcomes suggested that miR-592 may serve as a potential independent prognostic marker for RCC, which functioned similarly to several miRNAs in RCC, such as miR-154-5p,32 miR-23a-3p,33 and miR-663a.34
Next, we investigated the role of miR-592 on cell proliferation, migration, and invasion using 786-O and Caki-1 cells. The results showed that overexpression of miR-592 promotes the proliferation, migration, and invasion of RCC cell lines, while inhibition of miR-592 suppresses these cellular behaviors, compared with untreated cells. Thus, we speculate that miR-592 may serve as an oncogene in RCC. In colorectal cancer, miR-592 also plays an oncogenic role in tumorigenesis by targeting Forkhead Box O3A (FoxO3A).30 In gastric cancer, the ectopic expression of miR-592 promoted gastric cancer proliferation, migration, and invasion and facilitated tumorigenesis through the PI3K/AKT and MAPK/EPK signaling pathways by targeting Spry2.29 SPRY2 is a member of the signaling pathway-specific inhibition protein sprouty family, which has significant biological effects.35–37 For instance, downregulation of SPRY2 gene expression mediated by miR-21 promotes the proliferation and invasion of multiple myeloma cells.38 A previous study by Li et al have demonstrated SPRY2 was downregulated in RCC tissues and was associated with prognosis and inhibited cell proliferation and invasion in RCC cell lines.39 In our investigation, Targetscan predicted that SPRY2 was the direct target of miR-592 and luciferase reporter assays showed that SPRY2 3ʹ-UTR was the direct target of miR-592. According to previous study by Li et al39 and our present results, we speculate that miR-592 may promote tumor cell proliferation, migration, and invasion by targeting SPRY2. In the present study, these findings suggested that miR-592 may function as an oncogene and a potential prognostic biomarker in RCC. To date, we investigated the clinical significance of miR-592 and its potential cellular function in vitro, and it would be interesting to further investigate the detailed molecular mechanism of miR-592 in RCC.
Conclusion
Taken together, this study showed that miR-592 expression was upregulated in human RCC tissues and cell lines, and miR-592 directly targeted the expression of SPRY2 in RCC cells. Overexpression of miR-592 may promote proliferation, migration, and invasion of RCC cells by targeting SPRY2. Therefore, these data suggested that miR-592 may function as a novel oncogene that regulates RCC progression, which may provide valuable evidence for miR-592 as a prognostic biomarker and therapeutic target for RCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
3. Patel C, Ahmed A, Ellsworth P. Renal cell carcinoma: a reappraisal. Urol Nurs.2012;32:182–190. doi:10.7257/1053-816X.2012.32.4.182
4. Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913–924. doi:10.1016/j.eururo.2015.01.005
5. Shao N, Wan F, Abudurexiti M, et al. Causes of death and conditional survival of renal cell carcinoma. Front Oncol. 2019;9:591. doi:10.3389/fonc.2019.00591
6. Siska PJ, Beckermann KE, Rathmell WK, et al. Strategies to overcome therapeutic resistance in renal cell carcinoma. Urol Oncol. 2017;35:102–110. doi:10.1016/j.urolonc.2016.12.002
7. Petejova N, Martinek A. Renal cell carcinoma: review of etiology, pathophysiology and risk factors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160:183–194. doi:10.5507/bp.2015.050
8. Morales S, Monzo M, Navarro A. Epigenetic regulation mechanisms of microRNA expression. Biomol Concepts. 2017;8:203–212. doi:10.1515/bmc-2017-0024
9. Kim DY, Sung JH. Regulatory role of microRNAs in the proliferation and differentiation of adipose-derived stem cells. Histol Histopathol. 2017;32:1–10. doi:10.14670/HH-11-798
10. Zuo Z, Ye F, Liu Z, et al. MicroRNA-153 inhibits cell proliferation, migration, invasion and epithelial-mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp Ther Med. 2019;17:4693–4702. doi:10.3892/etm.2019.7470
11. Sun Y, Cao L, Lin JT, et al. Upregulated miRNA-1236-3p in osteosarcoma inhibits cell proliferation and induces apoptosis via targeting KLF8. Eur Rev Med Pharmacol Sci. 2019;23:6053–6061. doi:10.26355/eurrev_201907_18418
12. Berti FCB, Salviano-Silva A, Beckert HC, et al. From squamous intraepithelial lesions to cervical cancer: circulating microRNAs as potential biomarkers in cervical carcinogenesis. Biochim Biophys Acta Rev Cancer. 2019. doi:10.1016/j.bbcan.2019.08.001
13. Cui M, Wang H, Yao X, et al. Circulating MicroRNAs in cancer: potential and challenge. Front Genet. 2019;10:626. doi:10.3389/fgene.2019.00626
14. Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. doi:10.1615/CritRevEukaryotGeneExpr.v27.i3
15. Gao S, Chen J, Wang Y, et al. MiR-592 suppresses the development of glioma by regulating Rho-associated protein kinase. Neuroreport. 2018;29:1391–1399. doi:10.1097/WNR.0000000000001124
16. Wang W, Zhang H, Tang M, et al. MicroRNA-592 targets IGF-1R to suppress cellular proliferation, migration and invasion in hepatocellular carcinoma. Oncol Lett. 2017;13:3522–3528. doi:10.3892/ol.2017.5902
17. Liu M, Zhi Q, Wang W, et al. Up-regulation of miR-592 correlates with tumor progression and poor prognosis in patients with colorectal cancer. Biomed Pharmacother. 2015;69:214–220. doi:10.1016/j.biopha.2014.12.001
18. Fedorko M, Pacik D, Wasserbauer R, et al. MicroRNAs in the pathogenesis of renal cell carcinoma and their diagnostic and prognostic utility as cancer biomarkers. Int J Biol Markers. 2016;31:e26–e37. doi:10.5301/jbm.5000174
19. Dehghan F, Boozarpour S, Torabizadeh Z, et al. miR-21: a promising biomarker for the early detection of colon cancer. Onco Targets Ther. 2019;12:5601–5607. doi:10.2147/OTT.S199508
20. Huang D, Wang F, Wu W, et al. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting the AKT1 in melanoma. Cancer Biomark. 2019. doi:10.3233/CBM-190289
21. Peng P, Chen T, Wang Q, et al. Decreased miR-218-5p levels as a serum biomarker in bone metastasis of prostate cancer. Oncol Res Treat. 2019;42:165–185. doi:10.1159/000495473
22. Li B, Meng YQ, Li Z, et al. MiR-629-3p-induced downregulation of SFTPC promotes cell proliferation and predicts poor survival in lung adenocarcinoma. Artif Cells Nanomed Biotechnol. 2019;47:3286–3296. doi:10.1080/21691401.2019.1648283
23. Silva-Santos RM, Costa-Pinheiro P, Luis A, et al. MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer. 2013;109:2646–2653. doi:10.1038/bjc.2013.552
24. Huang QB, Ma X, Zhang X, et al. Down-regulated miR-30a in clear cell renal cell carcinoma correlated with tumor hematogenous metastasis by targeting angiogenesis-specific DLL4. PLoS One. 2013;8:e67294. doi:10.1371/journal.pone.0067294
25. He T, Chen P, Jin L, et al. miR6605p is associated with cell migration, invasion, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2018;17:2051–2060. doi:10.3892/mmr.2017.8052
26. Peng X, Pan X, Liu K, et al. miR-142-3p as a novel biomarker for predicting poor prognosis in renal cell carcinoma patients after surgery. Int J Biol Markers. 2019;34(3):302–308. doi:10.1177/1724600819866456
27. Hou W, Zhang H, Bai X, et al. Suppressive role of miR-592 in breast cancer by repressing TGF-beta2. Oncol Rep. 2017;38:3447–3454. doi:10.3892/or.2017.6029
28. Li Z, Li B, Niu L, et al. miR-592 functions as a tumor suppressor in human non-small cell lung cancer by targeting SOX9. Oncol Rep. 2017;37:297–304. doi:10.3892/or.2016.5275
29. He Y, Ge Y, Jiang M, et al. MiR-592 promotes gastric cancer proliferation, migration, and invasion through the PI3K/AKT and MAPK/ERK signaling pathways by targeting Spry2. Cell Physiol Biochem. 2018;47:1465–1481. doi:10.1159/000490839
30. Fu Q, Du Y, Yang C, et al. An oncogenic role of miR-592 in tumorigenesis of human colorectal cancer by targeting Forkhead Box O3A (FoxO3A). Expert Opin Ther Targets. 2016;20:771–782. doi:10.1080/14728222.2016.1181753
31. Lv Z, Rao P, Li W. MiR-592 represses FOXO3 expression and promotes the proliferation of prostate cancer cells. Int J Clin Exp Med. 2015;8:15246–15253.
32. Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;209:481–489. doi:10.1016/j.lfs.2018.08.044
33. Quan J, Pan X, Li Y, et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed Pharmacother. 2019;110:656–666. doi:10.1016/j.biopha.2018.11.065
34. Zhou L, Pan X, Li Z, et al. Oncogenic miR-663a is associated with cellular function and poor prognosis in renal cell carcinoma. Biomed Pharmacother. 2018;105:1155–1163. doi:10.1016/j.biopha.2018.05.082
35. Edwin F, Anderson K, Ying C, et al. Intermolecular interactions of sprouty proteins and their implications in development and disease. Mol Pharmacol. 2009;76:679–691. doi:10.1124/mol.109.055848
36. Lao DH, Chandramouli S, Yusoff P, et al. A Src homology 3-binding sequence on the C terminus of sprouty2 is necessary for inhibition of the Ras/ERK pathway downstream of fibroblast growth factor receptor stimulation. J Biol Chem. 2006;281:29993–30000. doi:10.1074/jbc.M604044200
37. Sayed D, Rane S, Lypowy J, et al. MicroRNA-21 targets sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi:10.1091/mbc.e08-02-0159
38. Wang JH, Zhou WW, Cheng ST, et al. Downregulation of sprouty homolog 2 by microRNA-21 inhibits proliferation, metastasis and invasion, however promotes the apoptosis of multiple myeloma cells. Mol Med Rep. 2015;12:1810–1816. doi:10.3892/mmr.2015.3567
39. Li P, Tao L, Yang J, et al. Sprouty2 is associated with prognosis and suppresses cell proliferation and invasion in renal cell carcinoma. Urology. 2013;82(253):e251–e257. doi:10.1016/j.urology.2013.02.051
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.