Back to Journals » OncoTargets and Therapy » Volume 14
AATF is Overexpressed in Human Head and Neck Squamous Cell Carcinoma and Regulates STAT3/Survivin Signaling
Authors Fu L, Jin Q, Dong Q, Li Q
Received 10 August 2021
Accepted for publication 22 October 2021
Published 9 November 2021 Volume 2021:14 Pages 5237—5248
DOI https://doi.org/10.2147/OTT.S333134
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Lin Fu,1,2 Quanxiu Jin,3 Qianze Dong,1,2 Qingchang Li1,2
1Department of Pathology, College of Basic Medical Science, China Medical University, Shenyang, People’s Republic of China; 2Department of Pathology, The First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China; 3Department of Breast Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, People’s Republic of China
Correspondence: Qingchang Li
Department of Pathology, College of Basic Medical Science, China Medical University, Shenyang, People’s Republic of China
Email [email protected]
Objective: Dysregulation of apoptosis antagonizing transcription factor (AATF) has been implicated in several cancers. However, its involvement in human head and neck squamous cell carcinoma (HNSCC) remains unclear. This study aimed to explore the expression pattern and biological roles of AATF in head and neck squamous cell carcinoma tissues and cell lines.
Methods: Immunohistochemistry was used to detect the level of AATF protein in 119 cases of HNSCC samples. CCK-8, colony formation, Annexin V/PI staining, Western blotting and RNA-sequencing were carried out to examine the change of proliferation, apoptosis and potential underlying mechanisms.
Results: Immunohistochemical staining showed that AATF was elevated in HNSCC, and high AATF level correlated with higher stage. The Cancer Genome Atlas (TCGA) and Oncomine data showed upregulated AATF expression in HNSCC compared with normal tissues. TCGA data also suggested that high AATF expression correlated with poor patient survival. Ectopic AATF expression upregulated the cell growth and colony formation ability in both FaDu and Detroit 562 cell lines, while AATF siRNA decreased the cell proliferation rate and colony numbers. AATF overexpression also decreased cisplatin sensitivity, downregulated cisplatin-induced apoptosis. Mechanistically, AATF overexpression upregulated survivin, while AATF knockdown downregulated survivin. RNA-sequencing (RNA-seq) and Gene Set Enrichment Analysis (GSEA) suggested that AATF knockdown decreased STAT3 signaling. Western blotting showed that AATF overexpression upregulated while AATF knockdown downregulated STAT3 phosphorylation. There was a positive correlation between AATF and survivin mRNA based on TCGA data analysis. Blockage of STAT3 signaling using inhibitor downregulated survivin expression and largely abolished the effects of AATF on survivin.
Conclusion: Our results showed that AATF was overexpressed in human HNSCC. AATF promoted cisplatin resistance and reduced apoptosis possibly through regulation of STAT3/survivin signaling. AATF could serve as a potential therapeutic target in HNSCC.
Keywords: AATF, survivin, STAT3, head and neck squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy and the leading causes of cancer-related death worldwide.1,2 Despite therapeutic advances, the prognosis of advanced stage HNSCC remains poor, with a 5-year survival rate less than 50%.3 The poor prognosis is in part due to the fact that numerous HNSCC develop drug resistance. Identifying the mechanisms underlying the HNSCC progression and seeking more effective targets for drug sensitivity are of great importance.
AATF is a transcriptional cofactor involved in the regulation of gene expression by connecting specific transcription factors to the general transcriptional machinery.4
AATF interacts with nuclear hormone receptors and enhances transactivation of several steroid hormone receptors, alone or in cooperation with histone acetyltransferase p300.5 In addition to nuclear hormone receptors, AATF has been shown to interact with several transcription factors, including the retinoblastoma protein (pRb), p65, and STAT3.6,7 Recent studies indicated AATF dysregulation in human cancers.8 AATF was reported to be amplified in neuroblastoma, the level of which is associated with poor patient prognosis.9 AATF could also promote multiple myeloma proliferation by affecting chromatin structure.10 However, the expression pattern of AATF and its biological roles in human HNSCC have not been explored. In this study, we examined the clinical significance, biological roles and underlying mechanisms of AATF in HNSCC tissues and cell lines.
Materials and Methods
Specimens
This study was approved by the ethics review board of China Medical University. Samples were collected from HNSCC patients who received surgical operations between April 2012 and December 2016. Of the 119 cases of HNSCC, 46 cases were from tongue, 60 were from larynx and 13 were from nasopharynx. Participants provided written informed consent and the study was performed according to the principles in the Declaration of Helsinki. The paraffin-embedded HNSCC specimens were obtained from the Pathology archives of the First Affiliated Hospital of China Medical University, which contain specimens no longer required to be maintained.
Immunohistochemistry
Sections (4-μm thick) were firstly treated with xylene for deparaffinization. Subsequently, sections were treated in graded ethanol for rehydration. Sections were treated with H2O2 solution to block the endogenous peroxidase. Heat antigen retrieval was carried out using citrate buffer (pH=6.0). Sections were washed with PBS and incubated with primary AATF antibody (1:200, Proteintech, USA) at 4°C overnight. ElivisionPlus HRP secondary antibody polymer (Maixin, Fuzhou, China) was used as the secondary antibody, and the sections were incubated at 37°C for 2 hours. Then, slides were stained by DABplus kit (Maixin, Fuzhou, China).
Immunostaining was evaluated by two pathologists from the pathology department in the First Affiliated Hospital of China Medical University. AATF was located at the nucleus of cancer cells. Staining intensity (0 negative/weak; 1 moderate; 2 strong) and percentage of AATF positive expression (1 (<25%); 2 (25–50%); 3 (50–75%); 4 (75–100%)) were scored separately. The final scores were obtained by multiplying the intensity scores with the percentage scores. Samples with scores <3 were considered as low expression.
Cell Culture, Transfection and Cisplatin Treatment
FaDu and Detroit 562 cell lines were obtained from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cell lines were maintained in PRMI-1640 with 10% fetal bovine serum (FBS) in an incubator with 5% CO2. pCMV6-hAATF plasmid (OriGene, USA) and pCMV6 empty vector (OriGene, USA) were transfected into HNSCC cells using Lipofectamine 3000 (Invitrogen, USA). AATF small interfering siRNA (GGACUUGGAUGAAGAAAUC) was purchased from Dharmacon (Horizon, Lafayette, CO, USA). Dharmafect1 reagent (Horizon, Lafayette, CO, USA) was used for siRNA transfection. Cisplatin was obtained from Selleck Chemicals (Houston, TX, USA). For cell viability assay, 2µM cisplatin was used to treat HNSCC cells for 24 and 48 hours. For apoptosis assay, 2µM cisplatin was used to treat HNSCC cells for 24 hours before detection.
Quantitative RT-PCR
TRIzol reagent (ThermoFisher, USA) was used to extract total RNA of HNSCC cells according to the manufacturer’s protocol. The concentration and purity (A260/280) of total RNA were detected by NanoDrop 2000 (ThermoFisher, USA). Then, cDNA was prepared via reverse transcription using PrimeScript RT Master Mix Kit (Takara, Dalian, China). PCR was performed using SYBR select master Mix Kit (Thermo, MA, USA). Relative expression of AATF was normalized to GAPDH using the 2−ΔΔct method. The primer sequences were as follows: AATF for 5′-CAGTCTACAGGAACCGCACAC-3′, AATF rev 5′-GTTCAAAGGCACCAAAACCCT-3′; GADPH for 5′-GAAATCCCATCACCATCTTCCAG-3′, GADPH rev 5′-GAGCCCCAGCCTTCTCCAT-3′.
Western Blotting
RIPA cell lysis was used to extract the total protein from HNSCC cells, and the concentration of total protein was determined by Bradford method. SDS-PAGE was used for protein separation, then the separated protein was transferred to PVDF membranes (Millipore, USA). The primary antibodies against AATF (1:1000, Proteintech, USA), survivin, p-STAT3, STAT3 (1:1000, Cell Signaling Technology, USA), Actin (1:2000, Santa Cruz, USA) were incubated at 4°C overnight. Then PVDF membranes were incubated with anti-mouse/rabbit peroxidase-conjugated secondary antibodies (1:2000, Cell Signaling Technology, USA) at 37°C for 2 hours. Membranes were visualized using ECL (Pierce, USA) and images were captured by DNR Bio-Imaging system (DNR, Israel).
CCK-8 and Colony Formation Assays
For CCK-8 assay, cells were plated in 96-well plates in medium containing 10% FBS at approximately 3000 cells per well 24h after transfection. 10 μL/per well of CCK-8 reagent (Cell Counting Kit-8; Dojindo, Kumamoto, Japan) was added and the cells were incubated at 37°C for another 2 hours. A microplate reader was used to detect the absorbance at 450 nm. For colony formation assay, HNSCC cells were seeded into 6-cm plates at the concentration of 1000 cells per plate. Then the plates were placed into an incubator for about 14 days. The cells were washed with PBS, fixed with 10% formaldehyde for 10 min and stained using Giemsa for 5 min. The number of cell colonies was counted under a microscope.
Annexin V/PI Apoptosis Staining
Cells were harvested and washed gently with PBS for three times. Then, the cells were suspended in a binding buffer. Next, 5 mg/mL propidium iodide (PI) and Annexin V/FITC antibody (BD Biosciences) were incubated with cells for 15 minutes in dark, then cell apoptosis was analyzed by flow cytometry.
RNA Sequencing
The RNA-seq experiments were performed by Novogene corporation (Beijing, China). The libraries were sequenced on an Illumina NovaSeq6000 platform. The sequence data was subjected to standard quality control. Heat map was generated with normalized data of RNA-seq analyses. Gene set enrichment analysis (GSEA) is performed using the software downloaded from GSEA website (http://software.broadinstitute.org).
Statistical Analysis
The software package SPSS 16.0 (SPSS, Chicago, IL) was used for data statistical analysis. A χ2 test was used to analyze the possible associations between AATF expression and clinicopathological parameters. Pearson’s correlation was used to analyze the correlation between AATF and survivin. Student’s t-test was used to compare the differences in gene expression, proliferation, apoptosis rate in groups. P<0.05 was considered statistically significant in all tests.
Results
AATF is Overexpressed in Human HNSCC
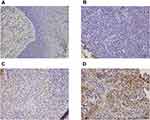
We collected 119 cases of HNSCC tissue and 12 cases of normal tissue, and used the immunohistochemistry method to examine the AATF protein levels. We found negative AATF staining in normal laryngeal squamous epithelium (Figure 1A). Increased AATF staining was found in 57 of 119 (47.8%) cases of HNSCC specimens (Figure 1B–D). As listed in Table 1, statistical significance was found between high AATF expression and T stage (T1-T2 vs T3-T4, p=0.0349), TNM stage (I+II vs III+IV p=0.0027). The associations between AATF levels and other parameters did not reach statistical significance.
 |
Table 1 Correlation Between the Clinicopathological Features and AATF Expression in HNSCC |
We analyzed the data from Oncomine database. Estilo Head-Neck, Talbot, and Ye Head-Neck of Oncomine datasets revealed that AATF mRNA was higher in tongue squamous cell carcinoma compared with normal tongue tissues (Figure 2A–C). Sengupta Head-Neck of Oncomine showed that AATF mRNA was higher in nasopharyngeal carcinoma compared with normal nasopharynx (Figure 2D). According to Ginos Head-Neck dataset of Oncomine, AATF mRNA was higher in HNSCC compared with buccal mucosa (Figure 2E). According to Peng Head-Neck dataset of Oncomine, AATF was higher in oral cavity squamous cell carcinoma compared with normal oral cavity (Figure 2F). In addition, we analyzed of the RNA-seq data of HNSCC from The Tumor Genome Atlas (TCGA). AATF mRNA in 502 cases of HNSCC was higher than that in normal tissues (n=44) (Figure 2G). We also analyzed AATF levels in 43 cases of paired HNSCC/normal tissues (Figure 2H). Statistical analysis showed that AATF showed significant higher expression in HNSCC tissues compared with corresponding normal tissues (Wilcoxon matched pairs signed rank test, p<0.001). Furthermore, analysis of TCGA also revealed that AATF mRNA was higher in HNSCC with higher pathological T stage (Figure 2I). Importantly, TCGA survival analysis showed that higher AATF levels significantly correlated with poor patient survival (p=0.0028) (Figure 2J). Taken together, these data indicated that AATF was upregulated in human HNSCC and correlated with malignant features. Figure 2 Continued.
AATF Promotes Proliferation in HNSCC Cell Lines
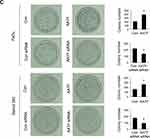
To explore the biological roles of AATF, we transfected both FaDu and Detroit 562 cell lines with the AATF plasmid and siRNA. RT-qPCR and Western blotting were used to validate of transfection efficiency, and the results are shown in Figure 3A. CCK8 assay was performed to assess the role of AATF on cell growth rate. Colony formation assay was performed to assess the role of AATF on colony formation capacity. As shown in Figure 3B, CCK-8 assays showed that AATF depletion downregulated the growth rate in FaDu and Detroit 562 cell lines while AATF overexpression showed the opposite results. As shown in Figure 3C, the number of colonies were decreased after AATF knockdown while increased after AATF overexpression. These results indicated that AATF promoted cell proliferation possibly through cell cycle control. Figure 3 Continued.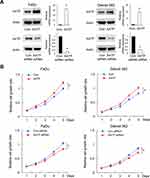
AATF Regulates Cisplatin Sensitivity and Apoptosis in HNSCC Cells
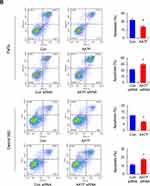
Next, whether AATF regulated cisplatin sensitivity of HNSCC cells was investigated. CCK8 assay was used to check the change of inhibition rate of cisplatin (2µM) in AATF overexpressed and depleted cells. According to CCK-8 assay results, the cisplatin inhibition rate was increased after AATF knockdown while was decreased after AATF overexpression when 24 and 48 h of treatment in both FaDu and Detroit 562 cell lines (Figure 4A). We also examined change of apoptosis rate after 24h of cisplatin treatment (2µM). Annexin V/PI assay showed that AATF overexpression decreased the cisplatin-induced apoptosis in FaDu and Detroit 562 cell lines, while AATF depletion upregulated cisplatin-induced apoptosis rate (Figure 4B). Figure 4 Continued.
AATF Regulates STAT3/Survivin Signaling
To explore the mechanism of AATF on cisplatin-induced apoptosis in HNSCC cells, we screened several apoptosis-related proteins and found that AATF overexpression increased while its knockdown decreased survivin expression, which functions as an anti-apoptosis protein (Figure 5C). We also analyzed RNA-sequencing data of 520 HNSCC cases from TCGA. The result showed that there were statistically significant positive correlations between AATF and survivin/BIRC5 (Pearson’s correlation, Figure 5B). To investigate the possible upstream mechanism of survivin upregulation, RNA-sequencing was carried out to profile mRNA change after AATF knockdown in FaDu cell line. GSEA analysis revealed significant enrichment for JAK/STAT3 signaling in control cells compared with AATF knockdown FaDu cells (Figure 5A). Western blotting demonstrated that ectopic AATF expression upregulated protein expressions of p-STAT3 in both FaDu and Detroit 562 cell lines (Figure 5C). STAT3 has been reported as an upstream signaling of survivin in various cancers. To further validate if survivin was regulated by AATF through STAT3 signaling, we blocked HNSCC cells using STAT3-specific inhibitor S31–201 (50 µM) and then tested the change of survivin. Western blot showed that STAT3 significantly reduced survivin expression in both cell lines. The upregulating effect of AATF on survivin was significantly decreased in cells treated with STAT3 inhibitor (Figure 5D). Figure 5 Continued.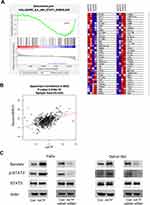
Discussion
Recent studies have implicated AATF functions as a cancer-related protein. To date, the expression pattern of AATF in head and neck squamous cell carcinomas remains unexplored. For the first time, our study showed AATF overexpression in head and neck squamous cell carcinoma tissues. AATF amplification has been reported in neuroblastoma and correlated with an adverse prognosis and reduced overall survival.9 AATF is significantly overexpressed in nonalcoholic fatty liver disease and hepatocellular carcinoma both in humans and mice.7 AATF elevation also contributes to poor prognosis of HCC patients.11
Our data demonstrated that AATF expression was elevated in 47.8% HNSCC tissues. Analysis of Oncomine and TCGA data also supported that fact of AATF upregulation in HNSCC. Higher AATF levels correlated with advanced TNM stage and T stage. Importantly, TCGA data analysis showed that AATF overexpression associated with poor patient survival. These results indicated that AATF signature might have clinical values in predicting malignant feature and providing prognostic information for HNSCC patients.
CCK-8 and colony formation assays demonstrated that AATF positively regulated HNSCC cell proliferation and colony formation ability. AATF is able to promote cell proliferation by interacting with histones and sustaining global histone acetylation in multiple myeloma cells.10,12,13 AATF knockdown decreased cell proliferation, migration, invasion, colony formation, and anchorage-dependent growth in HCC cell lines.7 Che-1 acts as a downstream effector of c-Myc and sustain proliferation in pre-B-cell acute lymphoblastic leukemia.14 AATF also promotes cell proliferation in a Kras-driven murine lung adenocarcinoma model.15 Our result was in accordance with previous reports, indicating AATF as a growth promoting protein in HNSCC.
Cisplatin is a standard and effective anticancer drug in multiple cancers including HNSCC. However, some patients exhibit drug resistance after treatment. Targeting resistance-related gene/protein may lower the effective dose, decrease side-effects, address drug resistance and improve overall survival outcomes, particularly for patients with difficult-to-treat cancers. Our results demonstrated that cisplatin sensitivity was suppressed, and the cisplatin-induced apoptosis level was decreased after AATF overexpression in HNSCC. While AATF depletion showed the opposite results, our data showed, for the first time, the involvement of AATF in chemo-sensitivity of HNSCC cells.
Next, we investigated the potential mechanism by which AATF regulates cisplatin-sensitivity. Our results showed that AATF positively regulated survivin protein expression in HNSCC cells. survivin is an anti-apoptotic mitochondrial protein that plays an important role during development of chemo-resistance. survivin expression above the median is associated with shorter overall survival in HNSCC, including for patients treated with chemotherapy or radiation.16 Survivin expression in HNSCC cells contributed to chemo-radioresistance, and its down-regulation increased anti-cancer effects of paclitaxel, cisplatin and radiation.17 In addition, TCGA data analysis revealed a positive association between AATF and survivin mRNA in HNSCC tissues.
Based on the above results, our data suggested the anti-apoptotic role of AATF in HNSCC cells might depend on its regulation of survivin.
To further explore the underlying mechanism by which AATF regulates survivin, we performed RNA-seq followed by GSEA analysis in FaDu cells transfected with control and AATF siRNA. Our data revealed enrichment for JAK-STAT3 signaling hallmark genes in control cells compared with AATF-depleted cells, suggesting AATF knockdown inhibited STAT3 signaling. Western blot further confirmed that AATF overexpression upregulated STAT3 phosphorylation, while AATF knockdown reduced STAT3 phosphorylation in HNSCC cell lines.
STAT3 is a transcription factor with a broad variety of biological functions.18 STAT3 is constitutively activated in a number of human cancers including HNSCC.19 STAT3 is activated by phosphorylation, which induces nuclear translocation, DNA binding, and mediates the expression of a variety of genes including survivin.20,21 STAT3/survivin axis has been reported in HNSCC. A previous report showed that AATF interacted with STAT3 to increase MCP-1 expression in HCC cell line.7 To validate if AATF could induce survivin through activation of STAT3, we treated AATF overexpressed HNSCC cells with STAT3 inhibitor S31–201. Inhibition of STAT3 largely abolished the effects of AATF on survivin, suggesting survivin act as a downstream target of STAT3 signaling in HNSCC cell lines. In addition, we noticed that STAT3 inhibitor slightly decreased endogenous AATF protein expression. This indicated AATF could also a downstream target of STAT3 signaling. Thus, there might be a positive feedback loop between AATF and STAT3, which needs further investigation. Together, our results indicated that AATF might modulate cisplatin sensitivity of HNSCC cells through regulation of STAT3/survivin pathway.
In conclusion, this study identified AATF was overexpressed in HNSCC and functioned as an oncoprotein. Our findings confirmed a link between AATF, STAT3 and cisplatin sensitivity of HNSCC cells, suggesting the therapeutic possibility of targeting AATF in HNSCC.
Acknowledgment
The study was supported by The National Natural Science Foundation of China (No. 81772468) to Qianze Dong, (No. 81702269) to Lin Fu and China Postdoctoral Science Foundation (2018M641736 and 2019T120222) to Lin Fu.
Disclosure
We declare that we have no conflict of interest.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29.
3. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi:10.1038/nrc2982
4. Srinivas AN, Suresh D, Mirshahi F, Santhekadur PK, Sanyal AJ, Kumar DP. Emerging roles of AATF: checkpoint signaling and beyond. J Cell Physiol. 2021;236(5):3383–3395. doi:10.1002/jcp.30141
5. Sharma M. Apoptosis-antagonizing transcription factor (AATF) gene silencing: role in induction of apoptosis and down-regulation of estrogen receptor in breast cancer cells. Biotechnol Lett. 2013;35(10):1561–1570. doi:10.1007/s10529-013-1257-8
6. Bruno T, Iezzi S, De Nicola F, et al. Che-1 activates XIAP expression in response to DNA damage. Cell Death Differ. 2008;15(3):515–520. doi:10.1038/sj.cdd.4402284
7. Kumar DP, Santhekadur PK, Seneshaw M, Mirshahi F, Uram-Tuculescu C, Sanyal AJ. A regulatory role of apoptosis antagonizing transcription factor in the pathogenesis of nonalcoholic fatty liver disease and hepatocellular carcinoma. Hepatology. 2019;69(4):1520–1534. doi:10.1002/hep.30346
8. Iezzi S, Fanciulli M. Discovering Che-1/AATF: a new attractive target for cancer therapy. Front Genet. 2015;6:141. doi:10.3389/fgene.2015.00141
9. Hopker K, Hagmann H, Khurshid S, et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 2012;31(20):3961–3975. doi:10.1038/emboj.2012.236
10. Bruno T, De Nicola F, Corleone G, et al. Che-1/AATF-induced transcriptionally active chromatin promotes cell proliferation in multiple myeloma. Blood Adv. 2020;4(22):5616–5630. doi:10.1182/bloodadvances.2020002566
11. Liu J, Lu J, Ma Z, Li W. A nomogram based on a three-gene signature derived from AATF coexpressed genes predicts overall survival of hepatocellular carcinoma patients. Biomed Res Int. 2020;2020:7310768.
12. Catena V, Bruno T, Iezzi S, et al. CK2-mediated phosphorylation of Che-1/AATF is required for its pro-proliferative activity. J Exp Clin Cancer Res. 2021;40(1). doi:10.1186/s13046-021-02038-x
13. Bruno T, De Nicola F, Goeman F, et al. Che-1/aatf-induced transcriptionally active chromatin promotes cell growth in multiple myeloma. Cancer Res. 2018;78(13). doi:10.1158/1538-7445.AM2018-350
14. Folgiero V, Sorino C, Pallocca M, et al. Che-1 is targeted by c-Myc to sustain proliferation in pre-B-cell acute lymphoblastic leukemia. EMBO Rep. 2018;19(3). doi:10.15252/embr.201744871
15. Welcker D, Jain M, Khurshid S, et al. AATF suppresses apoptosis, promotes proliferation and is critical for Kras-driven lung cancer. Oncogene. 2018;37(11):1503–1518. doi:10.1038/s41388-017-0054-6
16. Khan SA, Burke M, Zhu F, et al. Survivin expression and impact on head and neck cancer outcomes. Oral Oncol. 2021;112:105049.
17. Khan Z, Khan AA, Prasad GBKS, Khan N, Tiwari RP, Bisen PS. Growth inhibition and chemo-radiosensitization of head and neck squamous cell carcinoma (HNSCC) by survivin-siRNA lentivirus. Radiother Oncol. 2016;118(2):359–368. doi:10.1016/j.radonc.2015.12.007
18. Li B, Huang C. Regulation of EMT by STAT3 in gastrointestinal cancer (Review). Int J Oncol. 2017;50(3):753–767. doi:10.3892/ijo.2017.3846
19. Mali SB. Review of STAT3 (Signal Transducers and Activators of Transcription) in head and neck cancer. Oral Oncol. 2015;51(6):565–569. doi:10.1016/j.oraloncology.2015.03.004
20. Scheper MA, Nikitakis NG, Sauk JJ. Survivin is a downstream target and effector of sulindac-sensitive oncogenic Stat3 signalling in head and neck cancer. Int J Oral Maxillofac Surg. 2007;36(7):632–639. doi:10.1016/j.ijom.2007.04.003
21. Gritsko T, Williams A, Turkson J, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12(1):11–19. doi:10.1158/1078-0432.CCR-04-1752
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.