Back to Journals » Cancer Management and Research » Volume 14
A Technical Comparison of Human Papillomavirus Genotyping Assays from a Population-Based Cervical Cancer Screening in South Central Ethiopia
Authors Teka B , Gizaw M , Firdawoke E , Addissie A, Sisay TA, Schreckenberger C, Skof AS, Thies S, Mihret A , Kantelhardt EJ , Abebe T , Kaufmann AM
Received 1 February 2022
Accepted for publication 27 May 2022
Published 29 July 2022 Volume 2022:14 Pages 2253—2263
DOI https://doi.org/10.2147/CMAR.S360712
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Brhanu Teka,1,2 Muluken Gizaw,2– 4 Ededia Firdawoke,1 Adamu Addissie,3 Tesfamichael Awoke Sisay,3 Carola Schreckenberger,5 Anna Sophie Skof,5 Sarah Thies,5 Adane Mihret,1,6 Eva Johanna Kantelhardt,2,4 Tamrat Abebe,1 Andreas M Kaufmann5
1Department of Microbiology, Immunology and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia; 2Department of Gynaecology Martin-Luther-University, Halle-Wittenberg, Germany; 3School of Public Health, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia; 4Institute for Medical Epidemiology, Biometrics and Informatics, Martin-Luther-University, Halle-Wittenberg, Germany; 5Department of Gynecology, Charité – Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, 13353, Germany; 6Armauer Hansen Research Institute (AHRI), Addis Ababa, Ethiopia
Correspondence: Andreas M Kaufmann, Tel +49 30 450516499, Fax +4930 450-7 564958, Email [email protected]
Purpose: High-risk Human Papillomavirus (HPV) is the most important cause of cervical cancer. The highest burden of disease is seen in Low- and Low-Middle-Income Countries (LMIC). Several new HPV screening assays have been developed for high-risk HPV (hr-HPV) testing. We compared the performance and adequacy of three HPV genotyping assays on samples from a population of rural women in south-central Ethiopia.
Patients and Methods: One hundred and ten cervical swabs from rural women screened for HPV were assayed. HPV DNA was tested using MPG-Luminex Assay, Anyplex II HPV HR Detection, and EUROArray HPV. MPG-Luminex Assay was used as a reference method to compute the sensitivity and specificity of the two commercial assays in detecting hr-HPV infections.
Results: Of the 110 samples, MPG-Luminex Assay found 18.2% positive for the 14 hr-HPV and 7.3% for the probable hr-HPV genotypes. Anyplex™ II HPV HR Detection assay and EUROArray HPV Assay identified 21.82% and 12.7% samples, respectively, for the 14 hr-HPVs and both 7.3% for the probable hr-HPV genotypes (κ=0.734). Among the 14 hr-HPV genotypes, the genotype-specific agreement of the three HPV genotyping assays was moderate or better for HPV16, 31, 35, 39, 52, 56, 66 and 68. The aggregated sensitivity in detecting the 14 hr-HPV infections of Anyplex™ II HPV HR Detection and EUROArray HPV assays was high, 100% and 70%, respectively. The specificities of Anyplex™ II HPV HR Detection and EUROArray HPV were 95.6% and 100%, respectively.
Conclusion: The three evaluated assays showed similar analytical performance in the detection of hr-HPV infections and moderate or better concordance in HPV genotyping. This study is part of the ongoing cluster-randomized trial that has been registered in clinicaltrials.gov (NCT03281135) on September 13, 2017.
Keywords: analytical performance, HPV PCR test accuracy, HPV test complexity, HPV testing, LMIC
Introduction
Cervical cancer (CC) is the most common cancer in women in Sub-Saharan Africa.1 Its prevalence can be efficiently reduced by preventive HPV vaccination for certain hr-HPV genotypes and by having regular screenings to find clinically relevant pre-cancerous lesions followed by surgical removal.2 The World Health Organization (WHO) proposes a global strategy to eliminate cervical cancer through the 90-70-90 targets for 2030 (ie, vaccinating 90% of girls by age 15, screening 70% of women with high-performance tests by 35 and 45 years of age, and treating and/or manage 90% of women who tested positive for pre-cancer and invasive cancer). Thus, population-based screening remains important for both the vaccinated and more so the non-vaccinated population.3
In Ethiopia, the coverage of cytology-based cervical cancer screening is very low and it was estimated 1.6% and 0.4% in urban and rural areas, respectively.4 This is mainly due to the lack of gynecologists and pathologists needed for the procedure as well as awareness of women.5 In the current context of cervical cancer screening in Ethiopia for which the standard method is Visual Inspection with Acetic Acid (VIA), the acceptability and safety of the screening method are crucial parameters to enhance the coverage and improve the uptake by women.6
Detection of high-risk Human Papillomaviruses (hr-HPVs) in cervical smears has been shown to be more effective than cytology or VIA to detect CIN2+ dysplasia.7 Since hr-HPV infection is a necessary factor for CC development,2,8 a negative test for hr-HPV provides a relief from developing precancerous lesions for at least 5–10 years.9 In different studies, hr-HPV genotypes have been found to be associated with cervical and other carcinoma. For example, it has been indicated that more than 90% of anal and cervical cancers, about 70% of vaginal and vulvar cancers, 70% of oropharyngeal cancers and more than 60% of penile cancers are associated with hr-HPV infections.10 Among the HPV genotypes, the 14 HPV types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68) have been identified as high-risk (hr-HPV) for cervix cancer, but even among them the risk varies, and it is discussed to exclude HPV66 and 68.11
Numerous molecular diagnostic tools for the detection of HPVs have recently been developed and approved by several country-specific guidelines.12 HPV molecular testing has now become a screening tool to supplement other tests and/or as primary screening test alone. The current WHO cervical cancer screening test recommendation is to use HPV DNA as primary screening test either with or without triage to colposcopy or treatment to prevent cervical cancer among the general population of women.13 In addition, genotyping HPV tests are widely used in epidemiological studies, HPV surveillance, and vaccination impact monitoring.14 HPV genotyping tests for vaccine efficacy monitoring should cover the vaccine types, ie including the lr-HPV 6 and 11 present in the nonvalent vaccine, and have a high analytical sensitivity in order to find any infections irrespective of the presence of dysplasia. Furthermore, HPV genotyping is becoming crucial in risk stratification due to more or less carcinogenic types and type-specific persistence15 as risk for dysplasia development. Consequently, several commercial HPV genotyping assays are being successfully introduced for population-based HPV screening as well as for research purposes.
From the 2020 inventory of commercial molecular HPV tests, 254 distinct commercial HPV tests were identified in the global market. This represents a 31% increase in the number of distinct tests from 2015.14 Due to the diversity of detection methods, including HPV genotypes and targeted sequences between manufacturers, it is unavoidable that differences exist in genotype inclusivity and type-specific sensitivity or specificity among different HPV testing and genotyping assays.16 Another problem in selecting and introducing an HPV assay is that a significant number of these assays are without analytical and/or clinical evaluation according to international guidelines.17 Findings from recent studies reported that 60% of the HPV tests on the global market are still without a single peer-reviewed publication and 82% of tests lack any published analytical and/or clinical evaluation.14
Therefore, with a shift from cytology-based CC screening to HPV-based screening in many countries,18 a critical step is selecting an appropriate HPV test.17,19 Furthermore, accurate HPV genotyping methods are also required to assess the impact of HPV vaccination on virus prevalence. The HPV assay to be selected should be properly validated before use in terms of sensitivity (analytical and clinical), clinical accuracy, and reproducibility, as well it should offer rapid, affordable and preferably sample-to-answer solutions in diverse clinical and outpatient settings.20,21 In addition, high throughput capacity and point-of-care HPV tests are needed both with affordable prices and especially for LMIC where the main burden of disease and the least screening programs are established.
With this in mind, the aim of our comparison was to evaluate and compare the analytical performance of MPG-Luminex, Anyplex hr-HPV Detection and EUROArray HPV genotyping assays using samples collected from rural Ethiopia. We compared these three different genotyping HPV tests so as to establish an HPV DNA testing lab in Ethiopia, Addis Ababa University, as a national reference center for the purpose of population-based screening, epidemiological research, and vaccination efficacy surveillance. The study helped us to select tests with good performance in our context of LMIC both for screening and monitoring HPV vaccine efficacy. Furthermore, we have shown from our study that the complexity and degree of automation for all steps like the hands-on time, risk of contamination, and user-friendliness of the HPV assay were essential components to consider during validation and customization of assays in an LMIC context.
Materials and Methods
Study Samples
In a community-based follow-up study in Butajira, South Central Ethiopia, 110 cervical swabs were taken from women (aged 30 to 49) tested previously positive for hr-HPV in a previous screening round. Briefly, the parent study was a cluster-randomized trial that has been registered in clinical trial.gov (NCT03281135) and was conducted in Butajira Health and Demographic Surveillance Site (HDSS) of Addis Ababa University, Ethiopia. The study compared the uptake of VIA screening versus HPV DNA testing of self-collected cervicovaginal samples measuring participation, adherence to follow-up, and to determining the circulating HPV genotypes in the community following the same population after 6 and 24 months to evaluate the persistence, clearance, and reinfection rates of HPV genotypes. The study district is located 135 km south of the capital Addis Ababa.6,22 In this large, randomized control trial, 157 women were found to be hr-HPV positives from self-sampled specimen in a primary HPV screening round and invited for resampling to study the clearance and persistence of hr-HPV genotypes after six months of the primary screening. Of the hr-HPV positives in the primary screening, 110 women attended the follow-up and provided samples for HPV DNA testing after fulfilling the sample collection criteria.22 To evaluate the HPV genotyping assays of this study, we used these 110 samples collected by health care workers and compared the results from the three genotyping HPV tests. Samples were collected using the Cervex-Brush® (Rovers®, Oss, The Netherlands). It is a soft and flexible brush, which enables dual collection of ectocervical and endocervical samples, so all necessary cells can be collected in one movement. The Rovers Cervex-Brush device was rinsed immediately into PreservCyt® Solution filled ThinPrep vials (Hologic, Marlborough, USA) by pushing it onto the bottom of the vial 10 times, forcing the bristles apart followed by swirling the brush vigorously to further release material. The device was visually inspected to ensure that no material remained attached before it was discarded. The ThinPrep vial was labeled and stored at room temperature until shipment to the collaborator Laboratory for Gynecologic Tumor Immunology at Charité-Universitätsmedizin Berlin, Clinic for Gynecology, Germany, for DNA extraction and HPV genotyping.
DNA Extraction
DNA extraction was performed using an automated extraction method by Maxwell® 16 instrument and LEV Blood DNA kit (Promega, Madison, Wi, USA) following the manufacturer’s instructions. The DNA was eluted into 60 µL elution buffer and stored at −20°C until further use.
HPV Genotyping
HPV genotyping was carried out using the MPG-Luminex Assay following previous protocols,23 Anyplex™ II HPV HR (Seegene, Seoul, Korea) and EUROArray HPV (EUROIMMUN, Luebeck, Germany) according to the manufacturer’s recommended protocols for each assay.
Multiplexed Genotyping (MPG) by BSGP 5+/6+ PCR Followed by Luminex-Based Readout
This assay is a well-established assay proficient for HPV genotyping with high analytical sensitivity.24 MPG is an L1 gene-targeting PCR DNA test for HPV detection of 14 hr-HPV genotypes (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68), 4 probable hr-HPV (26, 53, 73 and 82) and 9 low-risk genotypes (HPV6, 11, 42, 43, 54, 57, 70, 72, and 90). The MPG assay also measures the cellular beta-globin gene of each sample, as a control for the adequate DNA amount. This genotyping assay was carried out generally as described by Schmitt et al.23 Broad-spectrum GP5+/6+ primers were used to amplify approximately 150 nucleotide long conserved target gene L1 ORF fragments. The final PCR reaction mix was 25 μL including 20 μL master mix and 5 μL DNA templates. Ten µL PCR products were hybridized with probe-conjugated Luminex beads, stained, and detected on a Bioplex 200 (Bio-Rad, Hercules, California).
EUROArray HPV
The EUROArray HPV assay (EUROIMMUN, Luebeck, Germany) is designed for the detection and genotyping of 30 human anogenital hr- and lr-HPV genotypes (HPV 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 61, 66, 68, 70, 72, 73, 81, 82, 89 (CP6108)) from DNA preparations of cervical smear samples. The assay was performed according to the manufacturers’ instructions.25 This assay combines multiplex polymerase chain reaction (PCR) amplification with oligonucleotide probe microarray for detection and genotyping of HPV DNA. HPV oncogenes E6 and E7 specific sequences are amplified and fluorescently labeled by means of a polymerase chain reaction (PCR) using a multiplex primer system. Fluorescently labelled amplicons bind to genotype-specific probes arranged in a microarray. The specific binding (hybridization) of a fluorescent PCR product to the corresponding oligonucleotide probe is detected using a special microarray scanner (EUROIMMUN). The EUROArray Scan software evaluates all spots, measures fluorescence signals, and generates the test results. The region of ubiquitous human Hsp90 gene serves as an endogenous control to verify DNA extraction and amplification adequacy. Moreover, it has a fully automated standardized evaluation, interpretation and archiving of the results through the integrated software.
Anyplex™ II HPV HR Detection
The Anyplex™ II HPV HR Detection (Seegene, Seoul, Korea) is a multiplex PCR assay with reporter detection designed for HPV genotyping. It can detect 14 hr-HPV genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68). It is a fully automated real-time PCR system. The PCR amplification is done using the CFX96 real-time thermocycler system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions.
The Anyplex assay utilizes TOCE (tagging oligonucleotide cleavage and extension) technology. The components of the assay are a dual priming oligonucleotide primer (DPO), Pitcher (a tagging oligonucleotide), and Catcher (a fluorescently labeled artificial template with a sequence complementary to the tagging portion of Pitcher). The DPO and Pitcher hybridize specifically on opposite sides of the target sequence of the HPV nucleic acid. The tagging portion of Pitcher is released during the DPO primer extension with Taq polymerase, which enables its hybridization to the capturing portion of Catcher. When “Duplex Catcher” (the tagging portion of Pitcher and the complementary Catcher sequence) is fully extended, it separates the reporter molecule from the quencher molecule, which results in a fluorescent signal. As an internal control (IC), the human housekeeping gene (beta-globin) is co-amplified simultaneously with the L1 gene sequences of the targeted HPV types in order to monitor nucleic acid isolation and check for possible PCR inhibition. The test result is generated automatically using Anyplex software.26
Statistical Analysis
The agreement of genotype results from three HPV genotyping assays was evaluated using the Fleiss’ kappa (κ) statistics. Agreement between the tests was assessed according to κ –values, where values in the range 0.81–1.00 indicate almost perfect agreement, 0.61–0.80 substantial, 0.41–0.60 moderate, 0.21–0.40 fair, 0.00–0.20 slight, and <0.00 poor agreement.27 Sensitivity and specificity were calculated using conventional contingency tables.
Results
The aim of this comparative study of HPV genotyping assays was to gain experience and produce comparative data for assay characteristics and complexity. There was a difference in categorization of carcinogenicity between the three evaluated HPV assays. Therefore, the carcinogenicity categorization in this comparison study was according to the protocols of each assay. HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68 were classified as hr-HPV and HPV26, 53, 73, 82 as probable hr-HPV genotypes and the other genotypes were considered as lr-HPVs. The spectrum of HPV genotypes detected in the three HPV genotyping assays is summarized in Table 1. The highlighted genotypes were matched in the assays and represent the 14 hr-HPVs to be compared.
 |
Table 1 Spectrum of HPV Genotypes in Three HPV Genotyping Assays |
Of the 110 samples, MPG-Luminex Assay, Anyplex™ II HPV HR Detection assay, and EUROArray HPV Assay detected 21.82%, 21.82%, and 16.36% hr-HPV genotype-positive samples for all hr-HPV types, respectively (κ=0.734) (Table 2). For the lr-HPV genotypes, the positivity rate was 5.45% in MPG-Luminex Assay and 7.27% in EUROArray HPV Assay that comprises 3 more genotypes (κ=0.237). The Anyplex™ II HPV HR Detection assay only includes hr-HPV genotypes (see Table 1).
 |
Table 2 HPV Positivity Result for the 110 Samples Tested with MPG-Luminex Assay, EUROArray HPV Assay, and Anyplex™ II HPV HR Detection Assay |
Concerning HPV genotypes, both hr- and lr-HPVs were detected in MPG-Luminex assay and EuroArray assay. In contrast, Anyplex™ II HPV HR Detection only includes 14 hr-HPV types. In all the 3 assays, the genotypes most often detected were HPV16, 35 and 52. HPV16 was detected in 6.4%, 3.6%, and 3.6% of the tested samples using MPG-Luminex Assay, EUROArray HPV Assay, and Anyplex™ II HPV HR Detection, respectively, while HPV35 was detected in 3.6%, 3.6%, and 4.5%, respectively. The other dominant genotype (HPV52) was detected in 6.4%, 2.7%, and 6.4% using MPG-Luminex Assay, EUROArray HPV Assay, and Anyplex™ II HPV HR Detection, respectively. Considering HPV82 that was included only in the MPG-Luminex Assay and the EUROArray HPV Assay, this genotype was the most predominantly detected by EUROArray HPV Assay (4.5%) and the second most prevalent in MPG-Luminex Assay (4.5%). Among the lr-HPVs, only HPV6 and HPV42 were commonly detected in both assays and HPV43 only by EUROArray.
The genotype-specific agreements of the three evaluated HPV genotyping assays are summarized in Table 3. For most of the genotypes, the assays showed moderate or better agreement. However, the level of discordance between the three assays was considerably high in the detection of HPVs 18 (κ=−0.003), 33 (κ=−0.003), 45 (κ=−0.003), 51 (κ=−0.012), 58 (κ=−0.003), 59 (κ=−0.003), and 73 (κ=−0.005). However, this analysis of agreement was limited by a very restricted number of positive samples for each genotype (Table 3). EUROArray HPV Assay did not detect any HPV51 and 68 infections compared with the other two assays. Anyplex™ II HPV HR Detection Assay detected HPVs 18, 33, 45, 58, and 59 which were not detected in MPG-Luminex and EUROArray HPV assays.
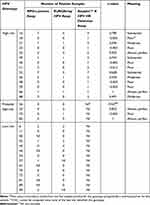 |
Table 3 Genotype-Specific Prevalence of HPV Genotypes in 110 Cervical Samples, by HPV Assay Used |
Next, we assessed the performance of the two commercial assays EUROArray HPV and Anyplex™ II HPV HR Detection Assay in the overall detection of the 14 hr-HPVs by using MPG-Luminex Assay as a reference method. This assay is based on the clinically validated GP5+/GP6+ PCR EIA that has been used as the gold standard comparator test for clinical validation of other tests. The MPG uses the GP5+/GP6+ primer set with consecutive probe-based genotyping via Luminex bead-based technology read out. It has been shown to have equal (relative) sensitivity and specificity like the EIA assay format.28
The aggregated sensitivity in detecting the 14 hr-HPV infections of EUROArray HPV and Anyplex™ II HPV HR Detection assays was high, 70% and 100%, respectively, while the specificities of EUROArray HPV and Anyplex™ II HPV HR Detection in the detection of the 14 hr-HPV infections were 100% and 95.6%, respectively, when compared with MPG-Luminex Assay. (Table 4).
 |
Table 4 Detection Performance for High-Risk HPVs of EUROArray HPV and Anyplex™ II HPV HR Detection Assays |
After the analysis of the HPV status among the three methods (comparison of only 14 hr-types detected in all three test), there were 11 cases (10% of the whole study population) which showed a discordant result for at least one of the HPV assays, resulting in overall HPV positivity or negativity (Table 5). The Anyplex™ II HPV HR Detection assay showed a discordant result for 5 of the 11 discordant results and indicated positive HPV status, which were negative in the other two assays. HPV18, 31, 51, and 56 were detected in the 5 discordant results by Anyplex™ II HPV HR Detection assay. For two samples, the MPG-Luminex assay displayed an HPV positivity (HPV51 and 52), whereas the EUROArray and Anyplex found no evidence of HPV positivity (see Table 5, sample ID 991374 and 991442). EUROArray HPV assay was discordantly negative for four samples, while the MPG-Luminex and Anyplex™ II HPV HR Detection assays were positive for those (Table 5). The four EUROArray HPV negative samples were positive for HPV16, 51, 52, 53, and 68 in MPG-Luminex assay, and for HPV 31, 52 and 68 in Anyplex™ II HPV HR Detection assay.
 |
Table 5 Discordant Cases for Hr-HPV Genotypes in the Comparison Among the Three HPV DNA Genotyping Assays |
Discussion
The current recommendation by WHO is to implement HPV-based cervical cancer screening. In addition, it is required to assess the impact of the HPV vaccination worldwide.29,30 For this purpose, only clinically validated tests should be used in clinical practice.31 Extended HPV genotyping is useful for population-level research purposes, where high analytical sensitivity, genotype-specific specificity and the ability to compare results between timepoints and populations are important.32 Therefore, it has become very important to optimize HPV diagnostic workflow in different populations and settings. In this study, we investigated the analytical performance of three HPV genotyping assays using samples collected from selected women who previously tested hr-HPV positive in a population-based follow-up study in rural Ethiopia.22
Although a good correlation of results was observed in both HPV positive and HPV negative samples, there were discordant results in positive samples among the three assays. The analytical performance of the different assays can be affected by many independent and interdependent factors including the assay intrinsic analytical sensitivity and specificity, or storage condition of cervical swab samples and DNA extraction method.33 However, in our case, because the samples were transported and processed under the same storage condition and the same DNA extraction method was used, the discrepancy of the results of the three assays was solidly influenced by other factors, such as inclusivity of HPV genotypes and type-specific sensitivity since the limits of detection inherent to an assay will determine by which sensitivity of each genotype is detected.32
Even though HPV16 and 18 are considered as the most important carcinogenic HPVs worldwide,34 there are also other HPV genotypes, which are categorized as carcinogenic, probably carcinogenic, or possibly carcinogenic.35 Therefore, due to their clinical significance, almost all available HPV genotyping assays can detect these high-risk group HPVs. For example, in our study, there were 14 overlapping hr-HPVs defined for detection between the three genotyping assays. Two of the evaluated assays include 18 hr-HPV and probable hr-HPV genotypes, while the third (Anyplex II HPV HR Detection) is restricted to 14 high-risk genotypes only. However, to this assay, a complementation assay exists testing for 18 additional probable high-risk and low-risk genotypes. Hence, this inclusivity differs between HPV genotyping assays and non-uniformity in the classification of carcinogenicity.
For population screening, the analytical sensitivity of HPV testing assays needs considerable attention because high analytical sensitivity does not guarantee acceptable diagnostic, ie, clinical sensitivity36 and specificity. Analytical sensitivity only represents the smallest amount of substance in a sample that can accurately be measured by an assay.37 It is clear that the analytical sensitivity differs among different HPV genotyping assays and may lead to controverted screening results between assays. Since the genotype-specific identification of HPVs with extended genotyping assays might be useful for test of cure, stratification of cancer risk, and to differentiate persistence from transient infections, genotype-specific validation of assays is crucial.31 Therefore, it is important to note that this study focused and compared the analytical sensitivity of three different HPV assays; however, this was not further supported by pathologic findings. Thus, a highly analytically sensitive test could detect a large number of clinically insignificant positive results. Therefore, for population screening, analytical sensitivity for all HPV genotypes included in each assay should be adjusted to the result of cervical pathological findings, and both clinical sensitivity and specificity are important for patient safety and must be considered in the context of using current and future HPV DNA tests.38
In this study, MPG-Luminex Assay was used as a reference genotyping test to determine the performance of the two commercial assays as it has high and equal (relative) sensitivity and specificity like the EIA assay format regarded as a gold standard.28 The sensitivity of the EUROArray HPV assay to detect the 14 hr-HPV genotypes was 70%, while Anyplex™ II HPV HR Detection Assay had an equal sensitivity (100%) with the reference assay. These differences can largely be explained by differences in the limits of detection of HPV genotypes in each assay. For example, among the less detected genotypes in EUROArray HPV assay in this study, HPV16 and 51, have different detection limits in the two evaluated assays (150 Vs 50 and 200 Vs 50 copies/PCR for EUROArray HPV and Anyplex™ II HPV HR Detection, respectively).32 The decreased detection of HPV52 by EuroArray was unexpected since the reported detection limit of both assays for this specific genotype seems similar (50 copies/PCR). However, this might be due to the low copy number of HPV52 in the specific sample tested so that it was potentially missed in one 5 µL sample taken for PCR in the EUROArray HPV detection. The low copy number of HPV52 can be explained by the weak signal strength measured by Anyplex™ II HPV HR Detection. Out of the four missed HPV52 HPV genotypes in EUROArray HPV detection, two were with weak signal strength (+) in the Anyplex™ II HPV HR Detection (data not shown).
Regarding the genotype-specific agreement of the three assays in our study, moderate and above moderate agreements were observed for more than half of the HPV genotypes evaluated. Substantial agreement was perceived between the three evaluated assays for HPV16, the most important carcinogenic genotype. HPV18, the other important genotype, was detected only in one woman by Anyplex™ II HPV HR Detection Assay but not by the other two assays. This could be due to the low copy number of HPV18 in the sample because it was detected with weak signal strength (+) in Anyplex™ II HPV HR Detection Assay. In our study, another genotype detection difference was observed in HPV68. HPV68 was detected more frequently in Anyplex™ II HPV HR Detection Assay than EUROArray HPV assay when compared to the reference assay. This is likely due to the variable efficiency of detection of HPV68 subtypes A and B. EuroArray is only able to amplify subtype HPV68A efficiently, while Anyplex II amplifies both HPV68A and 68B subtypes.32 Probes for both subtypes are also included in the MPG-Luminex assay.
Comparing the three assays with respect to the detection of the nonavalent HPV vaccine Gardasil 9 included hr-HPV genotypes, four of the genotypes (HPV18, 33, 45, and 58) out of the seven hr-HPV genotypes were only detected by Anyplex™ II HPV HR Detection Assay but not by the other two assays in the studied population. However, since the number of samples containing these types was too low (only single samples each positive) it cannot be concluded yet whether Anyplex™ II HPV HR Detection Assay would work better for vaccine surveillance. The major limitation of this study is its small sample size of HPV positive samples that was in part due to an unexpectedly high clearance rate in the hr-HPV positive sampled screening population. Therefore, some HPV genotypes had too low prevalence for calculation of assay sensitivity and specificity. Another limitation of this study was that the comparisons of the assays were performed based on a single test run. However, this study enabled us to establish the technical competence and initial prevalence information for planning of future trials.
In different parts of the world, the access to certain HPV assays varies. This mainly depends on the cost-effectiveness, infrastructure, complexity of the assay, and whether the assays fit with existing processes and equipment within an individual laboratory. Since newer assays are being developed and released regularly, it is important to determine relative performance and levels of agreement before introducing them for screening or diagnostic use in different settings. After our comparison study, we observed that the Anyplex™ II HPV HR Detection Assay is 100% sensitive and 95.6% specific compared to the reference assay in detecting the 14 hr-HPV genotypes. Furthermore, from our experience and observation during the study, the Anyplex™ II HPV HR Detection Assay was the easiest to handle with user-friendly workflow, requires less equipment, with few pipetting steps, a 96-well high through put capability, no post-PCR handling, with short hands-on time, choice of semi quantitation by the PCR program, automated result evaluation, and an open platform that can be easily used for add on tests. All these should be taken into account during HPV assay comparisons, choices and establishment in LMIC. Since most HPV testing systems were developed by and for HIC, it can be estimated that most currently available molecular HPV tests are too complex, eg, with large automated instruments needing expensive maintenance not readily available in LMIC, and/or costly for widespread use in LMIC. In our hands and as experienced in this study, the Anyplex™ II HPV HR Detection Assay was the simpler and more robust one in handling and instrumentation. This assay is also complemented by another accompanying test format (Anyplex II HPV28 Detection) that detects additional 14 probable and LR-HPV genotypes and can be used to achieve a more comprehensive picture for epidemiological investigations. Accordingly, we have now established Anyplex™ II HPV HR Detection Assay in the Ethiopian National HPV Reference Laboratory at the Department of Microbiology, Immunology & Parasitology, Tikur Anbessa Specialized Hospital, Addis Ababa University. This laboratory is in the process of accreditation to serve as a national HPV reference laboratory.
Conclusion
In conclusion, we evaluated two L1-and one E6/E7-targeting PCR DNA tests for the detection and differentiation of HPV genotypes. The three evaluated assays showed similar analytical performance as a screening tool for the 14 hr-HPV infections proposed by WHO for cervical cancer screening and moderate or better concordance in HPV genotyping. The complexity of assays is profoundly different and can also have an impact on assay choice. The sensitivity of the HPV assays compared was high; however, this was not further supported by colposcopy (or other triage tests) or any histologic findings to confirm that the detected hr-HPV positivity had any clinical significance in this study. Further research is required to confirm the clinical benefit in a LMIC setting that can be gained from the full genotyping offered by these assays. Thus, we have planned to conduct another study that addresses this issue.
Ethics Statement
This study was ethically approved by the Institutional Review Board of the College of Health Sciences, Addis Ababa University (057/17/SPH) and Martin Luther University, Halle Germany (2017-143). In addition, it was approved by the National Research Ethics Review Committee (NRERC) (SHE/RAAA/9.1/339/19/11) and a material transfer agreement was signed by both institutions to transfer samples to Germany. We certify that all participants gave informed consent and that the study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments.
Acknowledgments
We would like to thank all women who participated in our study and all community health workers of the Butajira district. A sincere gratitude also to Mrs Ursula Schiller (Charité—Universitätsmedizin Berlin, Germany) for facilitating and conducting laboratory work. Last but not least, the authors would also like to acknowledge Prof. Andreas Wienke (from Martin-Luther-University, Halle-Wittenberg, Germany) for his unreserved statistical advice during the analysis of this data.
Funding
This study was supported by Addis Ababa University, grant/award number: VPRTT/PY-041/2018 and BMZ/Else Kröner Fresenius-Stiftung; Else-Kröner-Fresenius Foundation. We acknowlege the financial support from the Open Access Publication Fund of Charite- Universitaetsmedizin Berlin and the German Research Foundation (DFG).
Disclosure
The authors declare that they have no conflicts of interest in relation to this work.
References
1. World Health Organization - International Agency for Research on Cancer. Globocan 2020 - Cervix uteri; 2020:2–3. Available from: https://gco.iarc.fr/today/data/factsheets/cancers/23-Cervix-uteri-fact-sheet.pdf.
2. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890–907. doi:10.1016/S0140-6736(07)61416-0
3. Roden RBS, Stern PL. Opportunities and challenges for human papillomavirus vaccination in cancer. Nature Rev Cancer. 2019;18(4):240–254. doi:10.1038/nrc.2018.13.Opportunities
4. Federal Democratic Republic of Ethiopia Ministry of Health. Guideline for cervical cancer prevention and control in Ethiopia; 2015.
5. Gizaw M, Teka B, Ruddies F, et al. Reasons for not attending cervical cancer screening and associated factors in Rural Ethiopia. Cancer Prev Res. 2020;13(7):593–599. doi:10.1158/1940-6207.CAPR-19-0485
6. Gizaw M, Teka B, Ruddies F, et al. Uptake of cervical cancer screening in Ethiopia by self-sampling HPV DNA compared to visual inspection with acetic acid: a cluster randomized trial. Cancer Prev Res. 2019;12(9):609–616. doi:10.1158/1940-6207.CAPR-19-0156
7. Chauhan AS, Prinja S, Srinivasan R, et al. Cost effectiveness of strategies for cervical cancer prevention in India. PLoS One. 2020;15(9):e0238291. doi:10.1371/journal.pone.0238291
8. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi:10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F
9. Dijkstra MG, van Zummeren M, Rozendaal L, et al. Safety of extending screening intervals beyond five years in cervical screening programmes with testing for high risk human papillomavirus: 14 year follow-up of population based randomised cohort in the Netherlands. BMJ. 2016;355:i4924. doi:10.1136/bmj.i4924
10. Arbyn M, de Sanjose S, Saraiya M, et al. EUROGIN 2011 roadmap on prevention and treatment of HPV-related disease. Int J Cancer. 2012;131(9):1969–1982. doi:10.1002/ijc.27650
11. Cuzick J, Adcock R, Wheeler CM. HPV genotype-specific risk for cervical cancer; 2021. Available from: www.HPVWorld.com.
12. Arbyn M, Simon M, Peeters E, et al. 2020 list of human papillomavirus assays suitable for primary cervical cancer screening. Clin Microbiol Infect. 2021;27(8):1083–1095. doi:10.1016/j.cmi.2021.04.031
13. World Health Organisation. WHO Guideline for Screening and Treatment of Cervical Pre-Cancer Lesions for Cervical Cancer Prevention.
14. Poljak M, Oštrbenk Valenčak A, Gimpelj Domjanič G, Xu L, Arbyn M. Commercially available molecular tests for human papillomaviruses: a global overview. Clin Microbiol Infect. 2020;26(9):1144–1150. doi:10.1016/j.cmi.2020.03.033
15. Elfgren K, Elfström KM, Naucler P, Arnheim-Dahlström L, Dillner J. Management of women with human papillomavirus persistence: long-term follow-up of a randomized clinical trial. Am J Obstet Gynecol. 2017;216(3):
16. Jones J, Powell NG, Tristram A, Fiander AN, Hibbitts S. Comparison of the PapilloCheck® DNA micro-array human papillomavirus detection assay with hybrid capture II and PCR-enzyme immunoassay using the GP5/6+ primer set. J Clin Virol. 2009;45(2):100–104. doi:10.1016/j.jcv.2009.02.013
17. Arbyn M, Snijders PJF, Meijer CJLM, et al. Which high-risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21(9):817–826. doi:10.1016/j.cmi.2015.04.015
18. Maver PJ, Poljak M. Primary HPV-based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;26(5):579–583. doi:10.1016/j.cmi.2019.09.006
19. de Sanjose S, Holme F. What is needed now for successful scale-up of screening? Papillomavirus Res. 2019;7:173–175. doi:10.1016/j.pvr.2019.04.011
20. Poljak M, Kocjan BJ, Oštrbenk A, Seme K. Commercially available molecular tests for human papillomaviruses (HPV): 2015 update. J Clin Virol. 2016;76:S3–S13. doi:10.1016/j.jcv.2015.10.023
21. Abreu ALP, Souza RP, Gimenes F, Consolaro MEL. A review of methods for detect human Papillomavirus infection. Virol J. 2012;9:1–9. doi:10.1186/1743-422X-9-262
22. Teka B, Gizaw M, Ruddies F, et al. Population-based human papillomavirus infection and genotype distribution among women in rural areas of South Central Ethiopia. Int J Cancer. 2021;148(3):723–730. doi:10.1002/ijc.33278
23. Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5 ϩ and GP6 ϩ primers. J Clin Microbiol. 2008;46(3):1050–1059. doi:10.1128/JCM.02227-07
24. Geraets DT, Cuschieri K, De Koning MNC, et al. Clinical evaluation of a GP5+/6+-based Luminex assay having full high-risk human papillomavirus genotyping capability and an internal control. J Clin Microbiol. 2014;52(11):3996–4002. doi:10.1128/JCM.01962-14
25. Cornall AM, Poljak M, Garland SM, et al. EUROarray human papillomavirus (HPV) assay is highly concordant with other commercial assays for detection of high-risk HPV genotypes in women with high grade cervical abnormalities. Eur J Clin Microbiol Infect Dis. 2016;35(6):1033–1036. doi:10.1007/s10096-016-2634-8
26. Hesselink AT, Sahli R, Berkhof J, et al. Clinical validation of AnyplexTM II HPV HR Detection according to the guidelines for HPV test requirements for cervical cancer screening. J Clin Virol. 2016;76:36–39. doi:10.1016/j.jcv.2016.01.009
27. Landis JR, Koch GG. Landis and Koch 1977_agreement of categorical data. Biometrics. 1977;33(1):159–174. doi:10.2307/2529310
28. Arbyn M, Hillemanns P. HPV assays validated for primary cervical cancer screening. HPV World. 2018;55:6–9.
29. Wentzensen N, Arbyn M, Berkhof J, et al. Eurogin 2016 Roadmap: how HPV knowledge is changing screening practice. Int J Cancer. 2017;140(10):2192–2200. doi:10.1002/ijc.30579
30. World Health Organization. Global Strategy Towards the Elimination of Cervical Cancer as a Public Health Problem. World Health Organization; 2019:1–24.
31. Arbyn M, Depuydt C, Benoy I, et al. VALGENT: a protocol for clinical validation of human papillomavirus assays. J Clin Virol. 2016;76(Suppl 1):S14–S21. doi:10.1016/j.jcv.2015.09.014
32. Cornall AM, Poljak M, Garland SM, et al. HPV genotype-specific concordance between EuroArray HPV, Anyplex II HPV28 and linear array HPV genotyping test in Australian cervical samples. Papillomavirus Res. 2017;4:79–84. doi:10.1016/j.pvr.2017.10.002
33. Donà MG, Benevolo M, Pimpinelli F, et al. Comparative evaluation of different DNA extraction methods for HPV genotyping by linear array and INNO-LiPA. J Med Virol. 2011;83(6):1042–1047. doi:10.1002/jmv.22088
34. Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17. doi:10.1128/CMR.16.1.1-17.2003
35. IARC WG. International Agency for Research on Cancer IARC monographs on the evaluation of carcinogenic risks to humans volume 90 human papillomaviruses; 2007. Available from: http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:Monographs+on+the+Evaluation+of+Carcinogenic+Risks+to+Humans+VOLUME+90+Human+Papillomaviruses#0.
36. Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front Public Heal. 2017;5:1–7. doi:10.3389/fpubh.2017.00307
37. Saah AJ, Hoover DR. “Sensitivity” and “specificity” reconsidered: the meaning of these terms in analytical and diagnostic settings. Ann Intern Med. 1997;126(1):91–94. doi:10.7326/0003-4819-126-1-199701010-00026
38. Kinney W, Stoler MH, Castle PE. Patient safety and the next generation of HPV DNA tests. Am J Clin Pathol. 2010;134(2):193–199. doi:10.1309/AJCPRI8XPQUEAA3K
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
