Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
A Simple Scoring System to Differentiate Bacterial from Viral Infections in Acute Exacerbations of COPD Requiring Hospitalization
Authors Ruiz-González A, Sáez-Huerta E, Martínez-Alonso M, Bernet-Sánchez A, Porcel JM
Received 13 January 2022
Accepted for publication 3 April 2022
Published 8 April 2022 Volume 2022:17 Pages 773—779
DOI https://doi.org/10.2147/COPD.S356950
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Agustín Ruiz-González,1 Eduardo Sáez-Huerta,1 Montserrat Martínez-Alonso,2 Albert Bernet-Sánchez,3 José M Porcel1
1Department of Internal Medicine, Arnau de Vilanova University Hospital, Biomedical Research Institute of Lleida (IRBLleida), University of Lleida, Lleida, Spain; 2Department of Statistics, Biomedical Research Institute of Lleida (IRBLleida), Lleida, Spain; 3Department of Microbiology, Arnau de Vilanova University Hospital, Lleida, Spain
Correspondence: José M Porcel, Department of Internal Medicine, Arnau de Vilanova University Hospital, IRBLleida, Avda Alcalde Rovira Roure 80, Lleida, 25198, Spain, Email [email protected]
Objective: Both bacteria and viruses may cause acute exacerbations of chronic obstructive pulmonary disease (AECOPD). The objective of this study was to identify readily available clinical parameters to discriminate between them.
Methods: During a winter period all consecutive patients with an AECOP who were hospitalized in a non-ICU general ward were prospectively enrolled. In addition to blood tests, cultures of spontaneous or induced sputum samples, and genome detection of respiratory viruses in nasopharyngeal swab samples using multiplex RT-PCR assays were obtained. Only patients with positive microbiological results (bacteria, virus, or both) were eventually included. Mixed infections (bacteria plus viruses) were categorized into the bacterial group due to therapeutic implications (ie, need for antibiotics). Demographic and routine clinical and analytical information was collected.
Results: A total of 127 AECOPD patients out of 213 initially evaluated met inclusion criteria and were classified as having bacterial (70, 55.1%) or viral (57, 44.9%) infection. Although no single variable was useful to identify bacteria, the combination of serum C-reactive protein > 70 mg/L (2 points), > 1 day of symptoms (1.5 points), and a blood neutrophil count > 9,500 x109/L (1 point) into a scoring system reached an AUC of 0.80 (95% CI=0.73– 0.88) for bacterial etiologies. With this model, scoring 0 or 1 point significantly reduced the probability of a bacterial infection (likelihood ratio negative of 0.2), whereas summing up 2.5 points or more increased it sufficiently to be clinically meaningful (likelihood ratio positive > 3.7). Viral infections resulted in fewer hospitalization days (78.9% of patients spent ≥ 3 days in hospital vs 95.7% of those with bacterial infections; P=0.008).
Conclusion: A simple and easy to obtain score system can help clinicians in the decision of prescribing antibiotics in AECOPD patients.
Keywords: COPD, respiratory infections, C-reactive protein
Introduction
An acute exacerbation of chronic obstructive pulmonary disease (AECOPD) is defined as an acute worsening of baseline respiratory symptoms that requires change in therapy. An AECOPD is associated with significant morbidity and impaired quality-of-life which are the primary drivers of hospital admissions. In this situation, guidelines recommend the use of antibiotics in patients with three cardinal symptoms (increased dyspnea, sputum volume, and sputum purulence), two cardinal symptoms of increased sputum purulence, plus either increased dyspnea or increased sputum volume, or the requirement for non-invasive or invasive mechanical ventilation.1 However, although only about 50% of AECOPD are caused by bacteria, most patients receive antibiotics with the risk of adverse side-effects and an increase in bacterial resistance.2
Previous studies have been conducted with the aim of identifying AECOPD caused by bacteria in an attempt to reduce antibiotic prescriptions. However, either the parameters described are not available in clinical practice or those that are available have controversial efficacy (eg, procalcitonin).3–8 The objective of the present study was to analyze whether there is a variable readily available at the patient’s bedside, either alone or in combination, capable of identifying bacterial infections in patients with AECOPD.
Methods
From December 2018 to March 2019 all consecutive patients admitted to the Emergency Room at the Arnau de Vilanova University Hospital with a main diagnosis of AECOPD who needed hospitalization in a regular-care ward were prospectively enrolled. The study was approved by the Ethics Committee of our institution (Comitè Ètic d’Investigació Clínica CEIC Hospital Universitari Arnau de Vilanova No. 1698) and all participants provided written informed consent.
The diagnosis of COPD was defined as a forced post-bronchodilator expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) ratio <0.7 in the adequate clinical context (ie, chronic and progressive symptoms of dyspnea, cough, and sputum production, with a history of exposure to risk factors). The level of obstruction (stage I–IV) followed the Global initiative for Chronic Obstructive Lung Disease (GOLD) guidelines.1 An AECOPD was defined as an increase in dyspnea, cough, and/or sputum volume or purulence in the last 2 weeks. Exclusion criteria were: 1) the use of corticosteroids and/or antibiotics during the 2-weeks pre-admission period, 2) an underlying malignancy or disease leading to immunosuppression, 3) non-infectious causes of AECOPD or other causes of dyspnea, such as heart failure, pulmonary embolism, or pneumonia, and 4) patients admitted to the ICU, a situation in which antibiotics are always indicated.
In addition to routine blood analyses, sputum and nasopharyngeal swab samples were obtained. Spontaneous or induced sputum samples were cultured using standard media. When patients were unable to spontaneously expectorate, which occurred in about one-third of the cases, sputum was induced by the inhalation of a 3% hypertonic saline solution (10–15 mL into a nebuliser cup) for approximately 10 minutes. If there were <10 epithelial cells and >25 neutrophils in each field under 10x magnification, the sputum specimen was considered adequate and was processed. A quantity of ≥105 cfu/mL was considered positive for bacterial growth. Nasopharyngeal swab samples were tested for genome detection of the following viruses by multiplex RT-PCR assay (LightMix®, TIB Molbiol, Berlin, Germany): Influenza virus (A and B), Respiratory Syncytial virus, Adenovirus, Enterovirus, Metapneumovirus, Bocavirus, Rhinovirus, Coronavirus (serotypes NL63, 229E, OC43, HKU1, MERS), and Parainfluenza virus (serotypes 1, 2, 3, and 4).9,10 Any positive result was considered a true viral infection. Only patients with positive microbiological results (either bacteria, virus, or both) were finally enrolled and none was included more than once.
In addition to the microbiological results, the following parameters were recorded: demographics (age, gender), smoking status, Charlson comorbidity index, degree of airway obstruction, long-term oxygen therapy, hospitalizations due to AECOPD during the previous year, use of inhaled corticosteroids, days of symptoms before admission, sputum purulence and Anthonisen criteria,11 vital signs, blood tests (neutrophil count, neutrophil/lymphocyte ratio, arterial blood gas analysis, fibrinogen, C-reactive protein [CRP], procalcitonin), and prognosis (days of hospitalization, need for non-invasive ventilation or ICU admission, and in-hospital mortality).
Since patients with mixed infections (bacterial and viral) benefit from antibiotic therapy, the “bacterial infection” group referred to those patients in whom either a bacteria or a mixed infection was identified, while the “viral infection” group applied to those in whom only a virus was recognized.
Statistical Analysis
Considering a ratio of bacterial-to-viral respiratory infections of approximately 1.2 in the hospitalized patients, to address our objectives we required 69 and 58 patients with bacterial and viral infections, respectively, to detect a standardized mean difference (effect size) of at least 0.5 units assuming respective type I and II errors of 0.05 and 0.20.
Data are presented as numbers (%) for categorical variables and as medians (interquartile range) for continuous variables. For between-group comparisons, the Pearson’s Chi-square, Student’s t, and Mann–Whitney U-tests were used as appropriate. For selecting those variables that discriminated between bacterial and viral groups of patients, the Boruta algorithm and a logistic regression model were employed.12 A scoring system to identify bacterial infection was constructed assigning points to the variables with the highest odds ratios (OR). The calibration and discriminative capacity of the final model were assessed using the Hosmer-Lemeshow test and the area under the ROC curve, respectively. A two-sided P<0.05 denoted statistical significance. Statistical analysis was performed using the R program.13
Results
An initial population of 213 patients with AECOPD were evaluated for the study, of whom 86 (40.3%) were excluded for different reasons (Figure 1). Thus, a total of 127 patients (median age 74.2 years; male sex 86.6%) were included in the study. This population consisted of 11% current smokers and had a high comorbidity (median Charlson index 5.4) and severe airflow obstruction (median FEV1 43%). Its baseline characteristics are shown in Table 1.
 |
Table 1 Baseline Characteristics of Study Population at Hospital Admission |
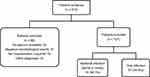 |
Figure 1 Flow diagram of the study population. Abbreviation: AECOPD, acute exacerbation of chronic obstructive pulmonary disease. |
The microorganisms identified in sputum samples are listed in Table 2. Bacterial, viral, or mixed (bacterial plus virus) infections were seen in 41 (32.2%), 57 (44.9%), and 29 (22.8%) patients, respectively. Bacterial and mixed infections were grouped together (70, 55.1%) for research purposes.
 |
Table 2 Microorganisms Identified in the Sputum of Patients with AECOPD |
As Table 1 shows, there were statistical differences between bacterial and viral groups in the following variables: FEV1, days of symptoms, purulence of sputum, respiratory rate, neutrophil count, neutrophil-to-lymphocyte ratio, fibrinogen, CRP, and procalcitonin. However, after application of Boruta’s algorithm, the variables with more weight for identifying bacteria were CRP (OR=8.17, 95% CI=3.50–20.8, P<0.001), days of symptoms (OR=7.61, 95% CI=1.99–35.8, P=0.005), and the blood neutrophil count (OR=2.84, 95% CI=1.18–7.16, p=0.022). A logistic regression model was performed after categorization of these variables, and a scoring system was developed as follows: serum CRP >70 mg/L (2 points), more than 1 day of symptoms (1.5 points), and a blood neutrophil count >9,500x109/L (1 point). Using this model, which had an AUC of 0.80 (95% CI=0.73–0.88), patients could be categorized as having low (22.2%), moderate (47.9%), or high (95.8%) risk of bacterial infection (Table 3). The operating characteristics of the model are shown in Table 4. Obtaining ≥3.5 points increased by about 60% the pre-test probability of a bacterial cause of AECOPD (likelihood ratio positive 18.7), whereas summing up one point or less decreased it by 30% (likelihood ratio negative 0.2). Overall, achieving 2.5 points was found to be clinically meaningful for labeling bacterial infections (likelihood ratio positive >3.7).
 |
Table 3 Scoring System and Its Calibration to Identify Bacterial Infections in Patients with AECOPD |
 |
Table 4 Operating Characteristics of the Scoring System to Identify AECOPD Due to Bacterial Infections |
During hospitalization, patients required noninvasive ventilation, admission to the ICU, or died in 11%, 3%, and 1.5%, respectively, and no differences were observed between bacterial and virus infection groups. However, 67 (95.7%) patients with bacterial infections spent 3 or more days in hospital, in comparison with 45 (78.9%) with virus infection alone (P=0.008).
Discussion
The present study shows that the combination of three simple and easy to obtain variables in the emergency room (CRP, days of symptoms, and blood neutrophil count) can accurately predict bacterial infection in AECOPD patients. A scoring system using these three variables (respective points of 2, 1.5, and 1) stratified patients according to the risk of having a bacterial infection from low (22.2%), moderate (47.9%), to high (95.8%). Obtaining 2.5 points argued for bacterial infection, whereas summing up 0 or 1 point argued against it.
GOLD guidelines recommend the use of antibiotics based on clinical features.1 The Anthonisen criteria, which include increased dyspnea, sputum volume, and sputum purulence, have been used for decades, but they are subjective and insufficiently accurate in predicting bacterial infection. The sputum color has been considered an acceptable indicator of bacterial infection in AECOPD patients. However, in a large study of 4,003 patients, the presence of green/yellow sputum (versus white color) was associated with bacteria with a sensitivity of 94.7%, though a specificity of only 15%.14
Many potential biomarkers have been tested in blood and/or sputum with the goal of predicting the bacterial origin of an AECOPD. Results have been varied and, oftentimes, suggested biomarkers are not widely available for routine use.3–8 Three examples on sputum specimens are worth mentioning. In a study of 145 patients with AECOPD, IL-1β in sputum samples was a good predictor of bacterial infection, with an AUC of 0.89 (95% CI=0.83–0.95).3 At a cut-off of 125 pg/mL, IL-1β showed a sensitivity and specificity of 90% and 80%, respectively. A second study of 124 patients with AECOPD showed that the combination of IL-8 and IL-10 levels in sputum was also useful to discriminate patients with bacterial infection from the non-infected ones and those with a viral infection (AUC=0.87, 95% CI=0.77–0.94).4 Finally, in a series of 90 AECOPD patients, sputum concentrations of neutrophil elastase identified bacterial infection with an AUC of 0.81 (95% CI=0.65–0.96).5 At the best cut-off of 3,034 ng/mL, test sensitivity and specificity was 77%. Unfortunately, sputum samples are not always available, and the preceding tests are limited to research settings.
The predictive value of several serum biomarkers is exemplified in the following investigations. A study of 63 patients showed that procalcitonin ≥0.25 ng/mL labelled bacterial infection with a sensitivity and specificity of 63% and 67%, respectively.6 Another study of 72 AECOPD patients was not able to demonstrate differences in the serum CRP and procalcitonin levels between bacteria-positive and bacteria-negative groups.7 In a small retrospective series of 45 hospitalized patients with AECOPD a neutrophil/lymphocyte ratio ≥4.5 had a sensitivity, specificity, and an AUC of 91%, 46%, and 0.75, respectively, to identify bacterial infections.8 However, the subjective clinical judgment in addition to culture results was incorporated to classify the patients.
Anticipating the bacterial etiology of an AECOPD may avoid the unnecessary use of antibiotics. A randomized controlled study of 653 AECOPD patients aimed to determine whether CRP point-of-care testing had an impact on antibiotic prescription. It was found that prescribing antibiotics when the CRP level was >40 mg/L decreased their use by 20% (from 77.4% to 57%), with no evidence of harm.15 Similarly, a meta-analysis of eight randomized trials totaling 1,062 AECOPD patients showed that indicating antibiotics only when the procalcitonin level was >0.25 ng/mL reduced antibiotic prescription (relative risk=0.56, 95% CI=0.43–0.73) without affecting clinical outcomes.16 However, no microbiological tests were performed on the patients.
The present study has strengths and limitations. Among the former, the study was prospective, only patients with positive microbiological results (bacteria, virus, or both) were included, and all blood and respiratory samples were obtained from antibiotic naïve patients, thus reducing the risk of false negative results. Our scoring system stratified patients into low, moderate, and high-risk of having bacterial infection. It could be argued that antibiotics may not be prescribed in patients who score 0 or 1 (likelihood ratio negative 0.2), while they would be fully indicated in those with a score of at least 2.5 (likelihood ratio positive >3.7).
Concerning study limitations, the research was conducted in a single center before the COVID-19 pandemic (we do not know if the results would change after the inclusion of SARS-Cov-2 virus), and critical patients directly admitted to the ICU were excluded (so, results cannot be extrapolated to such a population). Also, we should acknowledge the limited sensitivity of standard bacteriological cultures on sputum samples and, therefore, future studies using multiple PCR testing for typical and atypical bacteria are warranted.
In conclusion, bacterial infection in AECOPD patients can be easily predicted by a simple scoring system composed of serum CRP, days of symptoms, and blood neutrophil count. This information may assist clinicians in making decisions about the need to prescribe antibiotics.
Author Contributions
All authors meet criteria for authorship as recommended by the International Committee of Medical Journal Editors (ICMJE). All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Global Initiative for Chronic Obstructive Lung Disease (GOLD). Home page. Available from: http://www.goldcopd.org.
2. Celli BR, Barnes PJ. Exacerbations of chronic obstructive pulmonary disease [published correction appears. Eur Respir J. 2007;29(6):1224–1238. doi:10.1183/09031936.00109906
3. Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184(6):662–671. doi:10.1164/rccm.201104-0597OC
4. Dal Negro RW, Micheletto C, Tognella S, et al. A two-stage logistic model based on the measurement of pro-inflammatory cytokines in bronchial secretions for assessing bacterial, viral, and non-infectious origin of COPD exacerbations. COPD. 2005;2(1):7–16. doi:10.1081/copd-200050680
5. Thulborn SJ, Mistry V, Brightling CE, et al. Neutrophil elastase as a biomarker for bacterial infection in COPD. Respir Res. 2019;20(1):170. doi:10.1186/s12931-019-1145-4
6. Ergan B, Şahin AA, Topeli A. Serum procalcitonin as a biomarker for the prediction of bacterial exacerbation and mortality in severe COPD exacerbations requiring mechanical ventilation. Respiration. 2016;91(4):316–324. doi:10.1159/000445440
7. Chang CH, Tsao KC, Hu HC, et al. Procalcitonin and C-reactive protein cannot differentiate bacterial or viral infection in COPD exacerbation requiring emergency department visits. Int J Chron Obstruct Pulmon Dis. 2015;10:767–774. doi:10.2147/COPD.S76740
8. Van de Geijn GM, Denker S, Meuleman-van WV, et al. Evaluation of new laboratory tests to discriminate bacterial from nonbacterial chronic obstructive pulmonary disease exacerbations. Int J Lab Hematol. 2016;38(6):616–628. doi:10.1111/ijlh.12550
9. Hu HJ, Kim JY, Kwon HJ, et al. Performance evaluation of allplex respiratory panels 1, 2, and 3 for detection of respiratory viruses and influenza A virus subtypes. J Clin Microbiol. 2017;55(2):479–484. doi:10.1128/JCM.02045-16
10. Vandendriessche S, Padalko E, Wollants E, et al. Evaluation of the seegene allplex TM respiratory panel for diagnosis of acute respiratory tract infections. Acta Clin Belgica Int J Clin Lab Med. 2018;00:1–7. doi:10.1080/17843286.2018.1531605
11. Anthonisen NR, Manfreda J, Warren CP, et al. Antibiotic therapy in exacerbations of chronic obstructive pulmonary disease. Ann Intern Med. 1987;106(2):196–204. doi:10.7326/0003-4819-106-2-196
12. Kursa MB, Jankowski A, Rudnicki WR. Boruta - a system for feature selection. Fundam Inform. 2010;101(4):271–285. doi:10.3233/FI-2010-288
13. R Core Team (2020). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.R-project.org.
14. Miravitlles M, Kruesmann F, Haverstock D, et al. Sputum colour and bacteria in chronic bronchitis exacerbations: a pooled analysis. Eur Respir J. 2012;39(6):1354–1360. doi:10.1183/09031936.00042111
15. Butler CC, Gillespie D, White P, et al. C-reactive protein testing to guide antibiotic prescribing for COPD exacerbations. N Engl J Med. 2019;381(2):111–120. doi:10.1056/NEJMoa1803185
16. Mathioudakis AG, Chatzimavridou-Grigoriadou V, Corlateanu A, et al. Procalcitonin to guide antibiotic administration in COPD exacerbations: a meta-analysis. Eur Respir Rev. 2017;26(143):160073. doi:10.1183/16000617.0073-201
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
