Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
A Positive Feedback Loop of lncRNA HOXD-AS2 and SMYD3 Facilitates Hepatocellular Carcinoma Progression via the MEK/ERK Pathway
Authors Sun J , Li Y, Shi M, Tian H, Li J, Zhu K, Guo Y, Mu Y, Geng J, Li Z
Received 12 April 2023
Accepted for publication 20 July 2023
Published 27 July 2023 Volume 2023:10 Pages 1237—1256
DOI https://doi.org/10.2147/JHC.S416946
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Manal Hassan
Jin Sun,1– 3 Yingnan Li,1– 3 Mengjiao Shi,1,2 Hongwei Tian,1– 3 Jun Li,1– 3 Kai Zhu,1– 3 Ying Guo,1– 3 Yanhua Mu,1– 3 Jing Geng,1– 3 Zongfang Li1– 4
1National-Local Joint Engineering Research Center of Biodiagnostics and Biotherapy, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 2Shaanxi Provincial Clinical Research Center for Hepatic & Splenic Diseases, the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 3Center for Tumor and Immunology, the Precision Medical Institute, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 4Department of Geriatric General Surgery, the Second Affiliated Hospital of Xi’ an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China
Correspondence: Jing Geng; Zongfang Li, National & Local Joint Engineering Research Center of Biodiagnostics and Biotherapy, the second affiliated hospital, Xi’an Jiaotong University, No. 157 West 5th Road, Xi’an, 710004, People’s Republic of China, Tel +86-29-87679700 ; Tel/Fax +86-29-87679508, Email [email protected]; [email protected]
Purpose: HOX cluster-embedded long noncoding RNAs (HOX-lncRNAs) have been shown to be tightly related to hepatocellular carcinoma (HCC). However, the potential biological roles and underlying molecular mechanism of HOX-lncRNAs in HCC largely remains to be elucidated.
Methods: The expression signature of eighteen HOX-lncRNAs in HCC cell lines were measured by qRT-PCR. HOXD-AS2 expression and its clinical significance in HCC was investigated by bioinformatics analysis utilizing the TCGA data. Subcellular localization of HOXD‐AS2 in HCC cells was observed by RNA-FISH. Loss‑of‑function experiments in vitro and in vivo were conducted to probe the roles of HOXD-AS2 in HCC. Potential HOXD-AS2-controlled genes and signaling pathways were revealed by RNA-seq. Rescue experiments were performed to validate that SMYD3 mediates HOXD-AS2 promoting HCC progression. The positive feedback loop of HOXD-AS2 and SMYD3 was identified by luciferase reporter assay and ChIP-qPCR.
Results: HOXD-AS2 was dramatically elevated in HCC, and its up-regulation exhibited a positive association with aggressive clinical features (T stage, pathologic stage, histologic grade, AFP level, and vascular invasion) and unfavorable prognosis of HCC patients. HOXD-AS2 was distributed both in the nucleus and the cytoplasm of HCC cells. Knockdown of HOXD-AS2 restrained the proliferation, migration, invasion of HCC cells in vitro, as well as tumor growth in subcutaneous mouse model. Transcriptome analysis demonstrated that SMYD3 expression and activity of MEK/ERK pathway were impaired by silencing HOXD-AS2 in HCC cells. Rescue experiments revealed that SMYD3 as downstream target mediated oncogenic functions of HOXD-AS2 in HCC cells through altering the expression of cyclin B1, cyclin E1, MMP2 as well as the activity of MEK/ERK pathway. Additionally, HOXD-AS2 was uncovered to be positively regulated at transcriptional level by its downstream gene of SMYD3.
Conclusion: HOXD-AS2, a novel oncogenic HOX-lncRNA, facilitates HCC progression by forming a positive feedback loop with SMYD3 and activating the MEK/ERK pathway.
Keywords: hepatocellular carcinoma, HOXD-AS2, SMYD3, MEK/ERK signaling
Introduction
Hepatocellular carcinoma (HCC), as the major histologic category of primary liver cancer, is the seventh most prevalent neoplasm and the third-leading reason of cancer-associated mortality around the world.1 Despite substantial advancements have been achieved in HCC therapy, involving surgical resection, liver transplantation, chemotherapy, radiotherapy, especially systemic therapy, the mortality rate of HCC remains high due to its insidious nature and high rates of metastasis and recurrence.2,3 Consequently, there is an urgent demand to explicate the molecular mechanism behind HCC occurrence and development for offering efficient diagnostic biomarkers and therapeutic targets for HCC.
Class I homeobox genes (HOX genes), which represent the main subset of the homeobox family, are evolutionary conserved genes encoding transcription modulators that control the embryonic development in animal. In humans, 39 HOX genes are distributed in four clusters (HOXA, B, C, and D) and situated in chromosomal region of 7p15, 17q21.2, 12q13, and 2q31, correspondingly.4 Accumulating evidence shows that HOX genes were dys-regulated in a range of cancers including HCC and performed critical functions in the cancer development and progression.5,6 Interestingly, genome-wide transcriptome analyses revealed hundreds of sense or antisense long noncoding RNAs (lncRNAs) were transcribed from four HOX clusters, and these HOX cluster-embedded lncRNAs (HOX-lncRNAs) have been demonstrated to affect gene expression through both cis and trans mechanisms on HOX and non-HOX genes.7,8 For example, HOTAIR, a well-studied HOX-lncRNA with approximately 2.2 kilobases in length, is generated from antisense strand of HOXC gene cluster (between HOXC11 and HOXC12), and can impede transcription in trans across 40 kilobases of the HOXD locus through interplaying with polycomb repressive complex 2 (PRC2) and modulating H3K27me3 of HOXD locus.7 HOTTIP, another famous HOX-lncRNA with approximately 3.7 kilobases in length, is derived from the 5’ end of the HOXA cluster and can activate expression of posterior HOXA genes in cis through recruiting the WDR5/MLL complexes to the HOXA locus and driving the modification of H3K4 methylation.8
Noteworthily, some HOX-lncRNAs have been demonstrated to be abnormally expressed in various tumors including HCC, and closely linked with tumorigenesis as well as cancer progression as oncogenes or tumor suppressors.9 For instance, HOTAIR was uncovered to be overexpressed in HCC and was remarkably bound with poor tumor differentiation, metastasis as well as early recurrence.10 Biological function studies revealed that HOTAIR, as an oncogenic lncRNA, could promote HCC cell proliferation, metastasis and chemoresistance by multiple mechanisms, such as epigenetic suppression via DNA methylation and sponging microRNAs (miRNAs).11–13 Besides HOTAIR, several other HOX-lncRNAs, such as HOTTIP,14 HOXA11-AS,15 and HOXD-AS116 and so on, were also demonstrated to be linked with HCC development. Nonetheless, the potential biological functions and underlying molecular mechanism of HOX-lncRNAs in the occurrence and progression of HCC remain greatly ambiguous.
In the current research, the expression signature of eighteen referenced HOX-lncRNAs in five HCC cell lines and a normal hepatocyte cell line L-O2 were systematically inspected and HOXD Cluster Antisense RNA 2 (HOXD-AS2) was found to be the most significantly overexpressed HOX-lncRNA in HCC cells, and its expression status, clinical significance, biological functions as well as underlying mechanism in HCC progression were thoroughly characterized. This paper will help us acquire more profound awareness of the functional roles of HOX-lncRNAs in HCC development and provide promising new diagnostic biomarkers and therapeutic targets for HCC.
Materials and Methods
Human HCC Samples
A total of 30 samples of HCC tissues and matched neighboring noncancerous liver tissues were collected from patients who underwent surgical resection at Department of General Surgery, the Second Affiliated Hospital of Xi’an Jiaotong University between 2015 and 2017. All tissue specimens were pathologically verified confirmed and had not received radiotherapy or chemotherapy prior to collection. The tissues were frozen into liquid nitrogen as soon as dissected and then stored at −80°C. All patients received written informed consent. The study was permitted by the Ethics Committee of Xi’an Jiaotong University Health Science Center (No. 2019–1060), and all procedures were carefully handled to meet the guidelines of the Declaration of Helsinki.
Cell Culture
Five human HCC cell lines (Bel-7402, Bel-7404, SMMC-7721, HepG2, MHCC97H) and the normal hepatocyte cell line L-O2 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were authenticated by short tandem repeat (STR) profiling and were propagated in Dulbecco’s modified Eagle’s medium (Biological Industries, Israel) supplemented with 10% fetal bovine serum (Gibco, USA), penicillin–streptomycin (100 units/mL penicillin G sodium salt and 100 µg/mL streptomycin sulfate, Biological Industries, Israel) in a humidified atmosphere with 5% CO2 at 37°C.
Vector Construction, Small Interfering RNA (siRNA) Synthesis, and Transfection
For HOXD-AS2 knockdown, the lncRNA silencing experiment was performed using LncRNA Smart Silencer synthesized by RiboBio (Guangzhou, China), a mixture of three small interfering RNAs (siRNAs) and three antisense oligonucleotides (ASOs), and six target sequences of HOXD-AS2 are as follows: 5’-AACCUUCCUGAAAGAGAUGC-3’; 5’-GCAGAGACAAAGGAACUGCU-3’; 5’-CCAGGCUUCUUGGUGGCAUG-3’; 5’-CCAAGGAUUCAGCAAACAC-3’; 5’-CCCUCAUGGGCAUAGCCAU-3’; 5’-CCUGAAAGUAAAUGUCCUU-3’. For SMYD3 knockdown, the siRNAs specifically targeting SMYD3 (5’-AGCCUGAUUGAAGAUUUGATT-3’), and negative control siRNA (5’-UUCUCCGAACGUGUCACGUTT-3’) were also produced by RiboBio (Guangzhou, China). For SMYD3 overexpression, the open reading frame (ORF) of human SMYD3 was amplified and inserted into the mammalian expression vector pLVX-IRES-ZsGreen1 (Clontech, USA). To determine the promoter activity of HOXD-AS2 and SMYD3, DNA fragment encompassing the promoter region (−1500 to +100) were amplified and integrated into the pGL3-Basic vector (Promega, USA). All transfection experiments were conducted utilizing Lipofectamine® 3000 Reagent (Invitrogen, USA). The primers used for vector construction were given in Supplementary Table S1.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from tissues or cells utilizing RNAiso Plus reagent (TAKARA, China); nuclear and cytoplasmic RNA was extracted utilizing the PARIS™ Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. For qRT-PCR assay, cDNA template was generated by reverse transcription using HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, China) and qRT-PCR was conducted using SYBR™ Select Master Mix (Thermo Fisher Scientific, USA). All samples were normalized to beta-actin (β-actin) and the relative expression level was calculated according to the 2−ΔΔCt analysis methods. The primers used for qRT-PCR were listed in Supplementary Table S1.
RNA Fluorescent in situ Hybridization (RNA-FISH)
RNA-FISH assay was carried out utilizing RiboTM Fluorescent in Situ Hybridization Kit (RiboBio, China) in accordance with manufacturer’s protocol. Briefly, Bel-7402 cells were paved on the glass coverslips in a 24-well plate. When cells reached 60% confluency, they were fixed with paraformaldehyde (4%) and permeabilized with Triton X-100 (0.5%). The cells were then blocked with prehybridization buffer at 37°C for 40 min and incubated with 20 μM human HOXD-AS2 FISH Probe Mix (Cy3) (lnc1100245; RiboBio, China) at 37°C overnight. After washing by Hybridization Wash Buffer, cell nuclei were counterstained with DAPI. The glass coverslips carried cells were then placed on a glass slide and fluorescent images were observed using a fluorescence microscope (Leica DMI3000B, Germany). U6 and 18S rRNA were adopted as subcellular localization markers to indicate cytoplasmic and nuclear fraction, respectively, and FISH Probe Mix targeting U6 (lnc110101) and 18S (lnc110102) were also acquired from RiboBio (Guangzhou, China).
Cell Viability Assay
Cells (5×103 cells/well) were paved in 96-well plates and incubated with CCK-8 reagent (Dojindo, Japan) for 1 h 30 min at 37 °C. The optical density (OD) was collected at 450 nm by multifunctional microplate reader.
Colony Formation Assay
Cells were placed in 6-well plates at the density of 500 cells per well and cultured in complete medium for 7 days. When visible colonies appeared, the cells were fixed with 4% paraformaldehyde for 20 min. After staining with 0.1% crystal violet, the colonies were imaged and counted.
Cell Cycle and Apoptosis Analysis by Flow Cytometry
For cell cycle detection, the cells were fixed in 70% cold ethanol overnight at 4°C and then treated with DNA Content Quantitation Assay kit (Solarbio, China) containing propidium iodide (PI). The stained cells were then inspected by flow cytometry and the distributions of cells in distinct stages of the cell cycle were evaluated using FlowJo software (BD Biosciences, USA).
For apoptosis analysis, the cells were harvested and treated with Dead Cell Apoptosis Kit (Invitrogen, USA) consisting of Annexin V Alexa Fluor™ 488 and PI. The stained cells were then analyzed by flow cytometry and the proportion of apoptotic cells were measured by FlowJo software (BD Biosciences, USA).
Transwell Assay for Cell Migration and Invasion Detection
The capability of cell migration and invasion were assessed using matrigel-uncoated and coated 24-well transwell chambers with 8.0 μm transparent PET membrane (353097 and 354480, Corning, USA), respectively. A total of 5×104 cells were resuspended in 200 μL of serum-free medium and seeded in the upper chamber. The culture medium was supplemented with 10% FBS as a chemoattractant and added to the lower chamber. After 24 h (migration assay) or 48 h (invasion assay) of culture, the cells inside the chamber were wiped off and the cells outside the chamber were fixed with paraformaldehyde (4%). Subsequently, the migrated and invasive cells were stained with crystal violet and recorded under the microscope (100×) for counting.
Animal Experiments
Female BALB/c nude mice (four-week‐old) were purchased and raised in the Laboratory Animal Center of Xi’an Jiaotong University. Mice were randomly categorized into two groups (n = 5 for each group). 0.2 mL of HOXD‐AS2‐silenced or negative control Bel-7402 cells (containing 2 × 106 cells) were subcutaneously injected in the right flank of each nude mouse. The length and width of the tumors were checked every 7 days using vernier caliper and the volumes were evaluated according to the formula: volume = (length × width2)/2. At the end of the experiment, the mice were sacrificed, and the tumours were peeled off and weighed. All animal experiments were approved by the Animal Experimentation Ethics Committee of Xi’an Jiaotong University and were conducted in accordance with the institutional guidelines for care and use of laboratory animals.
Western Blot
Total proteins were prepared utilizing M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, USA) and histone was isolated by Histone Extraction Kit (Bestbio, China). Proteins were separated by SDS-PAGE on 4–20% gradient gels (GenScript, USA) and then transferred onto 0.22 μm PVDF membrane (Millipore, USA). After blocking with QuickBlock™ Western Blocking Buffer (Beyotime, China), the membranes were probed with the primary antibodies for 4°C overnight and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (1:10,000; AP307P, Millipore, USA) for 1 h at room temperature. The luminescence signal was visualized using the SuperSignal West pico chemiluminescent (Thermo Fisher Scientific, USA) and captured by digital chemiluminescence imager (Bio-Rad, USA). GAPDH or Histone H3 was applied as endogenous reference for total protein and histone. The primary antibodies used in this study were listed in Supplementary Table S2.
RNA-Sequencing (RNA-Seq) and Related Bioinformatics Analysis
Total RNA was extracted using RNAiso Plus reagent (TAKARA, Japan) and the integrity of the RNA was evaluated by the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNA-seq was executed by the Biomarker Biotechnology Corporation (Beijing, China) based on the Illumina system. Sequencing libraries were created utilizing NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was carried out on the cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumina, USA) and the libraries were then sequenced on an Illumina HiSeq Xten platform (Illumina, USA). The paired-end reads were filtered and mapped to the human reference genome (GRCh38/hg38) by TopHat2. Transcript expression levels were assessed by the value of FPKM (fragments per kilobase of transcript per million fragments mapped). The EBSeq algorithms were applied to screen out differentially expressed genes (DEGs) under the criteria of false discovery rate (FDR) < 0.05 and absolute value of log2-fold-change (log2FC) ≥0.6. GO and KEGG pathway enrichment was performed utilizing the DAVID online tool (https://david.ncifcrf.gov/) and statistically over-represented GO terms in the biological process (BP) and KEGG pathway were screened out by employing a p-value cutoff <0.05.
Dual-Luciferase Reporter Assay
Bel-7402 and SMMC-7721 cells were plated in 12-well plates and co-transfected with the constructed reporter vectors (1 µg), pRL-TK (0.2 µg), siRNA (50 nM) or expression vector (1 µg) utilizing Lipofectamine 3000 Reagent (Invitrogen, USA) for 48 h. Luciferase activity was assessed by Dual-Luciferase Reporter Assay System (Promega, USA), and the relative promoter activity was obtained by normalizing the Firefly luciferase activity with Renilla luciferase activity.
Chromatin Immunoprecipitation (ChIP) Assay
Bel-7402 cells were plated in 10 cm dish and transfected with SMYD3 overexpression or empty vector. After transfection for 48h, cells were cross-linked with 1% formaldehyde and ChIP assay was conducted utilizing SimpleChIP® Enzymatic Chromatin IP Kit (9003S, CST). Briefly, chromatin was fragmented (150 to 900 bp) using micrococcal nuclease and 2% of chromatin was kept as input. 8 µg of fragmented, cross-linked chromatin was immunoprecipitated with 2 µg of ChIP-grade SMYD3 antibody (GTX121945, GeneTex) and normal rabbit IgG (2729, CST). After reversal of the cross-link, immunoprecipitated DNA was isolated and quantified by qRT-PCR using gene-specific primers given in Supplementary Table S1. The percentage of input DNA were calculated according to the formula: Percent input = 2% × 2 (C[T] 2% input sample − C[T] IP sample).
Immunohistochemistry (IHC) Staining
Paraformaldehyde-fixed tissues were embedded in paraffin and then sliced into 4-μm thickness sections. After dewaxing, rehydration, and heat-mediated antigen retrieval, IHC staining was conducted using SP Rabbit & Mouse HRP Kit (DAB) (CW2069S, CWBIO, China) following the manufacturer’s protocol. Briefly, the sections were pretreated with endogenous peroxidase enzymes blocking buffer for 20 min and covered with normal goat serum for 25 min. Then, sections were labeled with anti-Ki67 antibody (1:200; sc-23900, Santa Cruz) and anti-PCNA antibody (1:500; 10205-2-AP, Proteintech) overnight at 4°C, followed by the treatment with biotinylated secondary antibody and HRP-labeled streptavidin. After visualizing with diaminobenzidine solution and counterstaining with hematoxylin, the images were obtained with a microscope (ZEISS, Axio Vert.A1).
Bioinformatics Analysis Based on TCGA Data
RNA-seq data and clinicopathological information of HCC cohort, comprising 374 HCC patients and 50 nonneoplastic tissues, were acquired from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The detailed clinical features of HCC patients are listed in Supplementary Table S3. The HTSeq-FPKM data was transformed into TPM (transcripts per million reads) data, and then normalized by log2 transformation for further analysis. All the analyses were performed using R (version 3.6.3). The expression level of HOXD-AS2 in HCC tissues was evaluated by the Mann–Whitney U-test and the median value was applied to classify the patients into high and low expression groups. The relationships between clinicopathologic features and HOXD-AS2 expression were analyzed by building the logistic regression model. Survival analysis was conducted with Kaplan‑Meier curves using survminer R package (v.0.4.9). The received operating characteristic (ROC) curve was drawn using pROC R package to estimate the diagnostic performance of HOXD-AS2. The expression relationship among HOXD-AS2 and its downstream genes were assessed by Pearson correlation analysis. Besides, the normalized expression matrix of GSE45436 and GSE101728 datasets was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and the expression levels of HOXD-AS2 were extracted to validate its expression trends in HCC tissues.
Statistical Analysis
All statistical analyses were carried out using SPSS 20.0. The data are expressed as the means ± standard deviation (SD). The differences between the two groups and multiple groups were determined by Student’s t-test and one-way analysis of variance (ANOVA), respectively. The expression of HOXD-AS2 in unpaired or paired HCC tissues was analyzed by performing Mann–Whitney U-test or Wilcoxon signed rank test, respectively. p<0.05 was taken as statistically significant.
Results
Expression Signature Analysis of Eighteen HOX-lncRNAs in HCC Cells by qRT-PCR
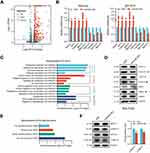
To survey the expression patterns of HOX-lncRNAs in HCC, we detected the expression status of 18 HOX-lncRNAs annotated by NCBI RefSeq database (see Figure 1A) in five HCC cell lines (Bel-7402, Bel-7404, MHCC97H, SMMC-7721, HepG2), as well as a normal hepatocyte cell line L-O2 using qRT-PCR. We found three lncRNAs (HOXB-AS1, HOXB-AS3 and HOXC-AS1) were not detected in HCC cells, and the expression status of the remaining 15 lncRNAs were shown in Figure 1B. Strikingly, HOXD-AS1 and HOXD-AS2, two lncRNAs both derived from HOXD locus, were significantly up-regulated in all five detected HCC cell lines. HOXA-AS2, HOXA-AS5, HOXA-AS6, HOXB-AS5, and HOXC-AS5 also displayed a high-expression trend in at least three HCC cell lines. Meanwhile, several lncRNAs, including HOXA-AS3, HOXA-AS4, HOXB-AS4, HOXC-AS2, and HOXC-AS3, were significantly down-regulated in HCC cells. Collectively, these results indicated that most HOX-lncRNAs exhibited abnormal expression patterns in HCC cells, implying that they may be related to HCC development.
Bioinformatics Analysis of HOXD-AS2 Expression and Its Clinical Significance in HCC Utilizing TCGA Data
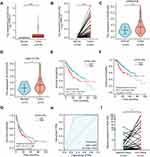
The above expression pattern analysis showed that HOXD-AS2 was the most remarkably overexpressed HOX-lncRNA in HCC cells (Figure 1B), indicating that HOXD-AS2 may exert crucial functions in HCC progression. To clarify the clinical significance of HOXD-AS2, we comprehensively analyzed the expression status of HOXD-AS2 and its clinical relevance in HCC utilizing TCGA data, including RNA-seq data and clinicopathological information. As indicated by Figure 2A and B, HOXD-AS2 was dramatically increased in both unpaired and paired HCC tissues when compared with normal liver tissues, and its up-regulation in HCC tissues was also validated in two GEO datasets (Figure 2C and D). Logistic regression analysis showed overexpressed HOXD-AS2 was markedly associated with gender, T stage, pathologic stage, histologic grade, AFP level, as well as vascular invasion (Table 1). Survival analysis indicated elevated HOXD-AS2 expression was notably correlated with worse overall survival (OS), disease-specific survival (DSS) as well as progression-free interval (PFI) (Figure 2E-G). ROC curves uncovered that the area under the curve (AUC) of HOXD-AS2 was 0.840, implying the HOXD-AS2 expression level possess a good diagnostic value for HCC (Figure 2H). Furthermore, the expression of HOXD-AS2 was verified in 30 pairs of HCC and neighboring non-neoplastic tissues, which we collected, and the qRT-PCR results exhibited HOXD-AS2 expression was elevated in tumor tissue (Figure 2I), which was in accordance with the bioinformatics analysis.
 |
Table 1 Relationship Between HOXD-AS2 Expression and Clinicopathologic Features (Logistic Regression) |
HOXD-AS2 is Present in Both the Nucleus and the Cytoplasm of HCC Cells
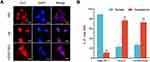
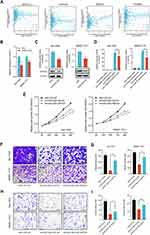
The biological functions of lncRNAs have been demonstrated to be intrinsically influenced by their subcellular localization. In general, nuclear lncRNAs can interact with chromatin modulators, transcription factors or ribonucleoproteins, whereas cytoplasmic lncRNAs are more likely to modulate mRNA stability or miRNA availability.17 To clarify the subcellular localization of HOXD-AS2 in HCC cells and provide clues into its functional and mechanistic study, we first detected the subcellular distribution of HOXD-AS2 in Bel-7402 cells by RNA-FISH and found that HOXD-AS2 transcripts could be probed in both the nucleus and the cytoplasm of Bel-7402 cells (Figure 3A); next, the cytoplasmic and nuclear RNA of Bel-7402 cells was separated, and the relative abundance of HOXD-AS2 transcripts was examined by qRT-PCR, and the results unveiled that HOXD-AS2 was more enriched in cytoplasm (Figure 3B). Collectively, our results suggested that HOXD-AS2 was distributed in both the nuclear and the cytoplasmic compartments of HCC cells, but more abundant in cytoplasm.
Downregulation of HOXD-AS2 Restrains Proliferation, Migration and Invasion Abilities of HCC Cells in vitro
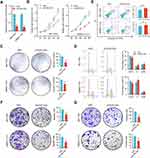
To address the biological function of HOXD-AS2 in HCC in vitro, Bel-7402 and SMMC-7721, two HCC cell lines highly expressed HOXD-AS2, were selected to perform loss-of-function experiment. As shown by Figure 4A, HOXD-AS2 could be successfully knocked down by LncRNA Smart Silencer. Subsequently, the impact of HOXD-AS2 knockdown on malignant biological behavior including proliferation, migration as well as invasion were investigated. As indicated by CCK-8 and colony formation assays, suppression of HOXD-AS2 dramatically restrained the proliferative activity of HCC cells (Figure 4B and C). To further determine whether the HOXD-AS2 knockdown affect the cell cycle and apoptosis, the flow cytometric detection was conducted to examine the proportion of cells in corresponding phases of cell cycle and undergo apoptosis after PI and/or annexin V staining, we revealed HOXD-AS2 silencing could trigger a cell cycle arrest at S and G2/M phases (Figure 4D). Interestingly, no significant effect on apoptosis was observed upon HOXD-AS2 knockdown (Figure 4E). Furthermore, the transwell assay disclosed HOXD-AS2 knockdown markedly impeded the ability of migration and invasion of HCC cells (Figure 4F and G). Altogether, our results disclosed that knockdown of HOXD-AS2 can attenuate malignant biological properties of HCC cells in vitro, indicating HOXD-AS2 exerts an oncogenic function in HCC.
Downregulation of HOXD-AS2 Represses the Growth of HCC Cells in Subcutaneous Xenograft Nude Mouse Model
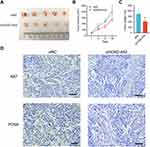
To further address the impact of HOXD-AS2 knockdown on tumor growth of HCC cell in vivo, we conducted in vivo experiments using subcutaneous xenograft nude mouse model established by Bel-7402 cells. As indicated in Figure 5A-C, the volume and weight of xenograft tumors were significantly suppressed after HOXD-AS2 knockdown when compared to the negative control group. IHC staining of the xenograft tissues uncovered that the expression levels of two proliferation markers (Ki67 and PCNA) were markedly diminished when HOXD-AS2 was knockdown (Figure 5D). Taken together, in vivo experiment demonstrates that downregulation of HOXD-AS2 can impede the growth capacity of HCC cells in vivo.
The Influence of HOXD-AS2 Knockdown on the Transcriptome of HCC Cells is Revealed by RNA-Seq
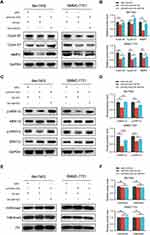
To survey which genes can be modulated by HOXD-AS2 in HCC cells, the transcriptome alternations of Bel-7402 cells upon HOXD-AS2 knockdown was determined by RNA-Seq. A total of 233 differentially expressed genes (DEGs) were screened out under the threshold of |log2(FC)| ≥ 0.6 and FDR < 0.05, including 190 up-regulated and 43 down-regulated genes, respectively (Figure 6A and Supplementary Table S4). It is noteworthy that the expression of some known oncogenes and tumor suppressors genes in HCC were disturbed after HOXD-AS2 knockdown, such as AKR1C3,18 TGFBR2,19 NOTCH2,20 SMYD3,21 SOCS1,22 ATF3,23 CDKN1C,24 BATF2,25 PRICKLE126 (Figure 6B). Next, to gain insight into probable biological functions and signaling pathways affected by HOXD-AS2 in HCC cells, GO and KEGG pathway enrichment analysis of DEGs were carried out to gain the enriched biological processes (BP) and signaling pathways utilizing the DAVID online tool (Supplementary Table S5 and Supplementary Table S6). As shown in Figure 6C, DEGs could be remarkably enriched in chromatin regulation, cell proliferation, migration, and angiogenesis. Combined with the cell phenotypes examined in this study, we further detected the effect of HOXD-AS2 knockdown on the expression of some crucial positive modulators that control cell cycle and metastasis progression using western blot, the results displayed the protein level of CYCLIN B1, CYCLIN E1 and MMP2 could be significantly decreased by HOXD-AS2 silencing (Figure 6D). In addition, we also found some vital cancer-associated pathways were also dramatically enriched by DEGs, like TNF and MAPK pathway (Figure 6E). The MEK/ERK cascade, the central constituent of MAPK pathway, has been well studied and shown to play vital functions in HCC growth and metastasis. Here, the impacts of HOXD-AS2 knockdown on the MEK/ERK pathway in HCC cells was validated and the results showed the levels of phosphorylated MEK1/2 (p-MEK1/2) and ERK1/2 (p-ERK1/2) was remarkably decreased upon HOXD-AS2 knockdown in Bel-7402 cells (Figure 6F), implying MEK/ERK signaling could be blocked by HOXD-AS2 knockdown in HCC cells.
SMYD3 is a Downstream Gene of HOXD-AS2 That Mediates the Oncogenic Roles of HOXD-AS2 in HCC Cells
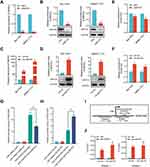
The above RNA-seq and qRT-PCR validation results disclosed four oncogenes (AKR1C3, NOTCH2, SMYD3, TGFBR2) that could be downregulated by HOXD-AS2 knockdown. Here, we further evaluated the expression correlations between these four oncogenes and HOXD-AS2 using TCGA data, and the results displayed only SMYD3 show an obvious positive correlation with HOXD-AS2 (Figure 7A), indicating SMYD3 might be the downstream target gene of HOXD-AS2. Next, the regulatory effect of HOXD-AS2 on SMYD3 was further verified by qRT-PCR and western blot, and we found HOXD-AS2 knockdown could decrease the expression of SMYD3 at both mRNA and protein levels in HCC cells (Figure 7B and C). The dual-luciferase reporter experiment revealed the promoter activity of SMYD3 could be repressed by the HOXD-AS2 knockdown, implying that HOXD-AS2 could govern the SMYD3 expression at the transcriptional level (Figure 7D). To determine whether SMYD3 mediated the regulation of HOXD-AS2 on biological behaviors of HCC cells, a series of rescue experiments was conducted. CCK8 assay revealed that overexpressing SMYD3 could restore the suppressive effect of HOXD-AS2 knockdown on the cell proliferative activity (Figure 7E). Transwell assays indicated that the prohibitive effects of HOXD-AS2 silencing on the capability of cell migration and invasion could be reversed through elevating SMYD3 (Figure 7F-I). Western blotting results indicated overexpression of SMYD3 could reverse the reduction of expression of CYCLIN B1, CYCLIN E1 and MMP2 as well as the inhibition of MEK/ERK pathway caused by HOXD-AS2 knockdown (Figure 8A-D). SMYD3, as a chromatin modifier, has been proved to promote gene transcription by increasing the level of H3K4me2 and H3K4me3.27 However, we did not observe the influence of HOXD-AS2 knockdown on the levels of H3K4me2 and H3K4me3 in HCC cells (Figure 8E and F). Taken together, these results imply that SMYD3 as downstream target mediates the oncogenic role of HOXD-AS2 in HCC cells.
HOXD-AS2 Can Be Positively Regulated by Its Downstream Gene of SMYD3 in HCC Cells
Previous research has illustrated that lncRNAs can form a positive feedback loop with their downstream genes to exert biological functions in HCC.28,29 SMYD3 is a key transcription modulator that can regulate gene expression as chromatin modifier or transcriptional coactivator.27 Thus, the positive correlation between HOXD-AS2 and SMYD3 expression drew us to suspect whether SMYD3 could modulate the HOXD-AS2 expression in HCC cells. Here, the expression of SMYD3 was successfully muted and overexpressed in HCC cells (Figure 9A-D), and SMYD3 silencing did indeed reduce the HOXD-AS2 expression, while up-regulation of SMYD3 could increase the HOXD-AS2 expression in HCC cells (Figure 9E and F). Dual-luciferase reporter assays further unveiled that SMYD3 knockdown could weaken the promoter activity of HOXD-AS2, while SMYD3 overexpression elevated the promoter activity of HOXD-AS2 in HCC cells (Figure 9G and H), implying that SMYD3 could control the HOXD-AS2 expression at the transcriptional level. SMYD3 is known to bind to specific motif (5′-CCCTCC-3′ or 5′-GGAGGG-3′) that exist in the promoter sequence of target genes.30 An examination of the HOXD-AS2 promoter sequence revealed that there was a GGAGGG motif at 82 to 87 bp downstream of the transcription start site (TSS) (Figure 9I), suggesting that SMYD3 may directly bind to the HOXD-AS2 promoter. The ChIP-qPCR analysis further demonstrated that DNA fragments containing SMYD3-binding motifs could be significantly enriched, while DNA fragments without SMYD3-binding motifs were not enriched (Figure 9J), suggesting that SMYD3 can bind directly to the HOXD-AS2 promoter. Collectively, these results demonstrate HOXD-AS2 can be positively regulated by its downstream gene of SMYD3 in HCC cells.
Discussion
Thanks to advances in deep transcriptome sequencing and large-scale genome sequencing projects, a batch of HOX-lncRNAs that transcribed from human four HOX loci have been uncovered, and some of which have been characterized to perform pivotal roles in tumorigenesis and tumor development. For HCC, despite several HOX-lncRNAs have been demonstrated to participate in tumor growth, metastasis, recurrence, and drug resistance through governing the expression of HOX gene or other genes,14–16,31 the biological roles of HOX-lncRNAs in HCC is still to be unearthed.
Herein, the expression patterns of 18 referenced HOX-lncRNAs in five HCC cell lines and a normal hepatocyte cell line were examined, and most of them were disclosed to be abnormally expressed in HCC cell lines, including several well-known HCC-related HOX-lncRNAs, such as HOXA11-AS (also known as HOXA-AS5), HOTTIP (also known as HOXA-AS6), HOXD-AS1.14–16 Intriguingly, we noticed that HOXD-AS2 was the most significantly up-regulated in HCC cells among all the detected lncRNAs, while whether it is associated with HCC currently remains unclear. Here, our integrative analysis of TCGA data first disclosed that up-regulation of HOXD-AS2 was positively associated with aggressive clinical features as well as poor prognosis of HCC patients and exhibited a good diagnostic performance for HCC, indicating HOXD-AS2 might be a potential diagnostic and prognostic biomarkers for HCC. Similarly, in gliomas, HOXD-AS2 was also demonstrated to be overexpressed in tumor tissue and positively related to glioma grade as well as worse prognosis.32,33 Interestingly, a recent study in gastric cancer (GC) demonstrated that HOXD-AS2 exhibit a low transcript abundance in GC tissues as well as a negative correlation with clinicopathological factors and worse overall survival,34 indicating that HOXD-AS2 may play distinct roles in different tumors.
Loss-of-function assay demonstrated knockdown of HOXD-AS2 could inhibit the capacities of proliferation, migration as well as invasion of HCC cells, indicating HOXD-AS2 exert carcinogenic roles in HCC. Similarly, studies in gliomas have shown that HOXD-AS2 also exerts its function as an oncogene and can accelerate glioma progression via facilitating cell proliferation, migration as well as invasion.32,33 A recent study revealed that HOXD-AS2 also exert a cancer-promoting role in the progression of non-small cell lung cancer (NSCLC) through modulating miR-3681-5p/DCP1A axis.35 However, in GC, HOXD-AS2 was found to play a tumor suppressive role, because overexpression of HOXD-AS2 could impede GC development through restraining the ability of proliferation, migration as well as invasion of GC cells.34 Furthermore, we uncovered HOXD-AS2 knockdown could arrest the cell cycle of HCC cells at S and G2/M phase, indicating HOXD-AS2 could regulate HCC cell proliferation through controlling cell cycle. Interestingly, in gliomas, HOXD‐AS2 was demonstrated to prohibit cell growth through triggering a G0/G1 cell cycle arrest,32 indicating that HOXD-AS2 can modulate cell cycle progression via different regulatory mechanisms in different tumor cells.
Subcellular localization analysis demonstrated that HOXD-AS2 was distributed both in the nuclear and the cytoplasmic compartments of HCC cells, hinting that HOXD-AS2 probably function in HCC through transcriptional or post-transcriptional regulatory mechanisms. In gliomas, HOXD-AS2 has been revealed to modulate the expression of mucosa-associated lymphoid tissue protein 1 (MALT1) at the post-transcriptional level by sponging miR-3681-5p.33 Whereas in GC, HOXD-AS2 was found to modulate the expression of its nearby gene HOXD8 in cis.34 Herein, we explored the potential HOXD-AS2-regulated genes in HCC cells by performing RNA-seq, 233 of DEGs were revealed to be aberrantly expressed after HOXD-AS2 knockdown in Bel-7402 cells. GO enrichment analysis showed DEGs could not only be enriched in the GO terms that related to cell proliferation and migration, but also enriched in some GO terms regarding chromatin regulation and angiogenesis, suggesting HOXD-AS2 might also exert its oncogenic function in HCC through affecting chromatin remodeling and angiogenesis. KEGG analysis revealed DEGs were associated with TNF and MAPK signaling pathway, and the impact of HOXD-AS2 on MAPK pathway in HCC cells was further validated by detecting alternations of MEK/ERK cascade, which is the central component of MAPK pathway. Interestingly, HOXD-AS1, another HOX-lncRNA in HOXD locus, has also been disclosed to activate the MEK/ERK pathway in HCC cells.16
SMYD3 (SET And MYND Domain Containing 3), a histone methyltransferase, has been demonstrated to be abnormally elevated in HCC and tightly related to worse clinical outcome of HCC patients, as well as promote proliferation and metastasis of HCC as oncogenic gene.21,36 In this study, SMYD3 was uncovered to be positively controlled by HOXD-AS2 in HCC cells, and its overexpression could partially abrogate the prohibitive effect of HOXD-AS2 knockdown on the malignant biological phenotype of HCC cells, indicating SMYD3, as downstream gene of HOXD-AS2, mediates the cancer-promoting effects of HOXD-AS2 in HCC. Moreover, we uncovered that up-regulation of SMYD3 could reversed the prohibitory effect of HOXD-AS2 knockdown on the activity of MEK/ERK pathway, hinting that SMYD3 as a downstream gene of HOXD-AS2, mediated the modulation of HOXD-AS2 on MEK/ERK cascade. In line with our findings, SMYD3 has been demonstrated to accelerate tumor progression by activating MEK/ERK signaling in other tumor types, such as colorectal cancer and lung adenocarcinoma.37,38 Interestingly, we found that knockdown of HOXD-AS2 did not alter the H3K4me2 and H3K4me3 modifications, suggesting that SMYD3 may mediate the regulation of HOXD-AS2 on HCC progression through other mechanisms. Although SMYD3 was first discovered as a histone methyltransferase, it has been shown that SMYD3 also can regulate the transcription of target genes as a transcription factor independently of its histone methylation activity.39 In addition, the regulatory effect of SMYD3 on H3K4 methylation has been disclosed to be varied in different cell types, for example, in breast cancer cell line MCF7, SMYD3 knockdown does not actually affect the global H3K4 methylation level.40
Finally, we revealed SMYD3 could positively modulate the expression of HOXD-AS2 via directly interacting with the promoter of HOXD-AS2 in HCC cells, indicating that HOXD-AS2 and SMYD3 participate in HCC development through forming a positive feedback loop. Our current research reveals for the first time that HOXD-AS2 can be involved in tumor progression by forming a regulatory circuit with transcription modulator.
Conclusion
In summary, this paper systematically disclosed the expression pattern of HOX-lncRNAs in HCC cells for the first time, and unveiled HOXD-AS2, a novel oncogenic HOX-lncRNA that up-regulated in HCC, accelerates HCC development via building a positive feedback loop with SMYD3 and activating the MEK/ERK pathway (Figure 10), suggesting that HOXD-AS2 might be a promising diagnostic and prognostic marker as well as therapeutic target for HCC.
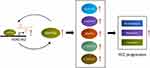 |
Figure 10 Schematic diagram of HOXD-AS2/SMYD3 reciprocal feedback loop facilitating HCC progression. |
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request by email ([email protected]).
Ethics Statement
The study was approved by the Ethics Committee of Xi’an Jiaotong University Health Science Center (No. 2019-1060). All experiments were conducted in accordance with the Declaration of Helsinki or the institutional guidelines for care and use of laboratory animals.
Acknowledgments
We would like to thank the other members of our research team for their help. We are also grateful to the TCGA and GEO database for the availability of the public data.
Funding
This study was supported by the National Natural Science Foundation of China (81502136 and 81802456), the Natural Science Foundation Research Program of Shaanxi Province of China (2023-JC-YB-750), the Science Research Foundation of the Second Affiliated Hospital of Xi’an Jiaotong University (2020YJ(ZYTS)546-09 and 2020YJ(ZYTS)645).
Disclosure
The authors declare no conflicts of interest.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Chakraborty E, Sarkar D. Emerging therapies for hepatocellular carcinoma (HCC). Cancers. 2022;14(11):2798. doi:10.3390/cancers14112798
3. Zhang H, Zhang W, Jiang L, Chen Y. Recent advances in systemic therapy for hepatocellular carcinoma. Biomark Res. 2022;10(1):3. doi:10.1186/s40364-021-00350-4
4. Luo Z, Rhie SK, Farnham PJ. The enigmatic HOX Genes: can we crack their code? Cancers. 2019;11(3):323. doi:10.3390/cancers11030323
5. Feng Y, Zhang T, Wang Y, et al. Homeobox genes in cancers: from carcinogenesis to recent therapeutic intervention. Front Oncol. 2021;11:770428. doi:10.3389/fonc.2021.770428
6. Jin Z, Sun D, Song M, et al. Comprehensive analysis of HOX family members as novel diagnostic and prognostic markers for hepatocellular carcinoma. J Oncol. 2022;2022:5758601. doi:10.1155/2022/5758601
7. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi:10.1016/j.cell.2007.05.022
8. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi:10.1038/nature09819
9. Nandwani A, Rathore S, Datta M. LncRNAs in cancer: regulatory and therapeutic implications. Cancer Lett. 2021;501:162–171. doi:10.1016/j.canlet.2020.11.048
10. Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11(3):1791–1798. doi:10.3892/ol.2016.4130
11. Cheng D, Deng J, Zhang B, et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. doi:10.1016/j.ebiom.2018.08.055
12. Guo Y, Liu B, Huang T, Qi X, Li S. HOTAIR modulates hepatocellular carcinoma progression by activating FUT8/core-fucosylated Hsp90/MUC1/STAT3 feedback loop via JAK1/STAT3 cascade. Dig Liver Dis. 2023;55(1):113–122. doi:10.1016/j.dld.2022.04.009
13. Duan Y, Chen J, Yang Y, et al. LncRNA HOTAIR contributes Taxol-resistance of hepatocellular carcinoma cells via activating AKT phosphorylation by down-regulating miR-34a. Biosci Rep. 2020;40(7):BSR20201627. doi:10.1042/BSR20201627
14. Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi:10.1002/hep.26740
15. Guo JC, Yang YJ, Zheng JF, et al. Silencing of long noncoding RNA HOXA11-AS inhibits the Wnt signaling pathway via the upregulation of HOXA11 and thereby inhibits the proliferation, invasion, and self-renewal of hepatocellular carcinoma stem cells. Exp Mol Med. 2019;51(11):1–20. doi:10.1038/s12276-019-0328-x
16. Sun J, Guo Y, Bie B, et al. Silencing of long noncoding RNA HOXD-AS1 inhibits proliferation, cell cycle progression, migration and invasion of hepatocellular carcinoma cells through MEK/ERK pathway. J Cell Biochem. 2020;121(1):443–457. doi:10.1002/jcb.29206
17. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220(2):e202009045. doi:10.1083/jcb.202009045
18. Zhou Q, Tian W, Jiang Z, et al. A positive feedback loop of AKR1C3-mediated activation of NF-kappaB and STAT3 facilitates proliferation and metastasis in hepatocellular Carcinoma. Cancer Res. 2021;81(5):1361–1374. doi:10.1158/0008-5472.CAN-20-2480
19. Chen YL, Xu QP, Guo F, Guan WH. MicroRNA-302d downregulates TGFBR2 expression and promotes hepatocellular carcinoma growth and invasion. Exp Ther Med. 2017;13(2):681–687. doi:10.3892/etm.2016.3970
20. Huang G, Jiang H, Lin Y, et al. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis. 2018;9(5):487. doi:10.1038/s41419-018-0554-5
21. Zhang H, Zheng Z, Zhang R, et al. SMYD3 promotes hepatocellular carcinoma progression by methylating S1PR1 promoters. Cell Death Dis. 2021;12(8):731. doi:10.1038/s41419-021-04009-8
22. Ding J, Xu K, Sun S, et al. SOCS1 blocks G1-S transition in hepatocellular carcinoma by reducing the stability of the CyclinD1/CDK4 complex in the nucleus. Aging. 2020;12(4):3962–3975. doi:10.18632/aging.102865
23. Chen C, Ge C, Liu Z, et al. ATF3 inhibits the tumorigenesis and progression of hepatocellular carcinoma cells via upregulation of CYR61 expression. J Exp Clin Cancer Res. 2018;37(1):263. doi:10.1186/s13046-018-0919-8
24. Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi:10.1038/onc.2008.178
25. Ma H, Liang X, Chen Y, et al. Decreased expression of BATF2 is associated with a poor prognosis in hepatocellular carcinoma. Int J Cancer. 2011;128(4):771–777. doi:10.1002/ijc.25407
26. Chan DW, Chan CY, Yam JW, Ching YP, Ng IO. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology. 2006;131(4):1218–1227. doi:10.1053/j.gastro.2006.07.020
27. Bernard BJ, Nigam N, Burkitt K, Saloura V. SMYD3: a regulator of epigenetic and signaling pathways in cancer. Clin Epigenetics. 2021;13(1):45. doi:10.1186/s13148-021-01021-9
28. Chen F, Bai G, Li Y, Feng Y, Wang L. A positive feedback loop of long noncoding RNA CCAT2 and FOXM1 promotes hepatocellular carcinoma growth. Am J Cancer Res. 2017;7(7):1423–1434.
29. Lan T, Yuan K, Yan X, et al. LncRNA SNHG10 Facilitates Hepatocarcinogenesis and Metastasis by Modulating Its Homolog SCARNA13 via a Positive Feedback Loop. Cancer Res. 2019;79(13):3220–3234. doi:10.1158/0008-5472.CAN-18-4044
30. Kim JM, Kim K, Schmidt T, et al. Cooperation between SMYD3 and PC4 drives a distinct transcriptional program in cancer cells. Nucleic Acids Res. 2015;43(18):8868–8883. doi:10.1093/nar/gkv874
31. Tang X, Zhang W, Ye Y, et al. LncRNA HOTAIR Contributes to Sorafenib Resistance through Suppressing miR-217 in Hepatic Carcinoma. Biomed Res Int. 2020;2020:9515071. doi:10.1155/2020/9515071
32. Qi Y, Wang Z, Wu F, et al. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression. J Cell Biochem. 2018;120(5):8343–8351. doi:10.1002/jcb.28117
33. Zhong X, Cai Y. Long non-coding RNA (lncRNA) HOXD-AS2 promotes glioblastoma cell proliferation, migration and invasion by regulating the miR-3681-5p/MALT1 signaling pathway. Bioengineered. 2021;12(2):9113–9127. doi:10.1080/21655979.2021.1977104
34. Yao L, Ye PC, Tan W, et al. Decreased expression of the long non-coding RNA HOXD-AS2 promotes gastric cancer progression by targeting HOXD8 and activating PI3K/Akt signaling pathway. World J Gastrointest Oncol. 2020;12(11):1237–1254. doi:10.4251/wjgo.v12.i11.1237
35. Zhang Y, Ma H. LncRNA HOXD-AS2 regulates miR-3681-5p/DCP1A axis to promote the progression of non-small cell lung cancer. J Thorac Dis. 2023;15(3):1289–1301. doi:10.21037/jtd-23-153
36. Wang Y, Xie BH, Lin WH, et al. Amplification of SMYD3 promotes tumorigenicity and intrahepatic metastasis of hepatocellular carcinoma via upregulation of CDK2 and MMP2. Oncogene. 2019;38(25):4948–4961. doi:10.1038/s41388-019-0766-x
37. Peserico A, Germani A, Sanese P, et al. A SMYD3 Small-Molecule Inhibitor Impairing Cancer Cell Growth. J Cell Physiol. 2015;230(10):2447–2460. doi:10.1002/jcp.24975
38. Mazur PK, Reynoird N, Khatri P, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510(7504):283–287. doi:10.1038/nature13320
39. Bottino C, Peserico A, Simone C, Caretti G. SMYD3: an Oncogenic Driver Targeting Epigenetic Regulation and Signaling Pathways. Cancers. 2020;12(1):142. doi:10.3390/cancers12010142
40. Van Aller GS, Reynoird N, Barbash O, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7(4):340–343. doi:10.4161/epi.19506
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.