Back to Journals » International Journal of General Medicine » Volume 14
A Novel DNA Damage Repair-Related Gene Signature for Predicting Glioma Prognosis
Authors Zhan J, Wu S, Zhao X, Jing J
Received 11 October 2021
Accepted for publication 6 December 2021
Published 21 December 2021 Volume 2021:14 Pages 10083—10101
DOI https://doi.org/10.2147/IJGM.S343839
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jiaoyang Zhan,1 Shuang Wu,2 Xu Zhao,3 Jingjing Jing4– 6
1Department of Anorectal Surgery, the First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China; 2College of Computer Science and Technology, Changchun Normal University, Changchun, Jilin, People’s Republic of China; 3Mathematical Computer Teaching and Research Office, Liaoning Vocational College of Medicine, Shenyang, Liaoning, People’s Republic of China; 4Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China; 5Key Laboratory of Cancer Etiology and Prevention in Liaoning Education Department, the First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China; 6Key Laboratory of GI Cancer Etiology and Prevention in Liaoning Province, the First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China
Correspondence: Jingjing Jing; Xu Zhao Email [email protected]; [email protected]; [email protected]
Background: Glioma is one of the most prevalent tumors in the central nervous system of adults and shows a poor prognosis. This study aimed to develop a DNA damage repair (DDR)-related gene signature to evaluate the prognosis of glioma patients.
Methods: Differentially expressed genes (DEGs) were extracted based on 276 DDR genes. Then, a gene signature was developed for the survival prediction in glioma patients by means of univariate, multivariate Cox, and least absolute shrinkage and selector operation (Lasso) analyses. After analyzing the clinical parameters, a nomogram was constructed and assessed. A total of 693 gliomas from the Chinese Glioma Genome Atlas (CGGA) were used for external validation. In addition, we used glioma tumor tissues for qPCR experiment to verify.
Results: A 12-DDR-related gene signature was identified from the 75 DEGs to stratify the survival risk of glioma patients. The overall survival of high-risk group was significantly shorter than that of low-risk group (P < 0.001). Besides, according to the risk score assessment, patients in high- or low-risk group also had significant correlations with clinicopathological parameters, including age (P < 0.01), grade (P < 0.001), IDH status (P < 0.001) and 1p19q codeletion status (P < 0.001). The nomogram provided favorable C-index and calibration plots. The C-index of training set and verification set was 0.761 and 0.746, respectively, and the calibration curve also showed that both training set and verification set were close to the standard curve. The qPCR results showed that there were significant differences in the expression of some typical DDR-related genes in tumor tissues and paracancer tissues (P(WEE1)=0.0002, P(RECQL)=0.0117, P(RPA1)=0.021, P(RRM1)=0.0035, P(PARP4)=0.0006, P(ELOA)=0.0023).
Conclusion: Our study developed a novel 12 DDR-related gene signature as a practical prognostic predictor for glioma patients. A nomogram combining the signature and clinical parameters was established as an individual clinical prediction tool.
Keywords: DNA damage repair-related gene signature, glioma, prognosis, nomogram, qPCR
Introduction
As one of the most prevalent tumors in central nervous system of adults, glioma accounts for approximately 50% of all the malignant brain tumors.1 The prognosis of patients with glioma is very poor, although a variety of therapies have been available, including surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy.2,3 Treatment resistance and tumor recurrence lead to treatment failure and adverse outcomes.4,5 Therefore, glioma is still one of the most challenging malignant tumors in the world. It is urgent to find a group of effective biomarkers for the diagnosis and treatment of glioma.
In all kinds of biological activities, DNA damage is inevitable DNA damage repair (DDR) plays an essential role in maintaining the integrity and stability of genome. DNA damage can lead to activation of oncogenes and inactivation of tumor suppressor genes, which is an important mechanism of cancer development.6,7 Dysregulation of DDR genes could also enhance the resistance of cancer cells to chemotherapy. DDR-related genes have been reported to act as therapeutic targets in prostate cancer and ovarian cancer.8,9 In addition, other studies have pointed out that different polymorphisms of DDR genes can influence each other and jointly affect the susceptibility of glioma.10,11 For example, two genotypes of DDR gene XRCC1, Arg/Gln(AG) and Gln/Gln(GC), are significant risk factors for glioma, and people with Gln(G) allele have a more than three times higher risk of glioma than normal people.12 Inhibition of DDR gene expression can improve the sensitivity of glioma to chemotherapy.13
Recently, plenty of gene signatures have become biomarkers for guiding treatment in various cancers.14–16 Multigene prognostic sets can predict the tumor prognosis more accurately than a single gene, which enable more effective treatment of cancer patients.17,18 Identifying the hallmark gene signature is important for clinical application and precision medicine of cancers. However, there is a lack of research on the prognosis of glioma with combined biomarkers. Therefore, it is of considerable importance to identify prognostic signatures of glioma based on the DDR-related gene expression profiles. In this study, we aimed to evaluate the association between DDR-related gene expression and molecular characteristics and survival of glioma, thus to establish a novel DDR-related gene signature for predicting glioma prognosis.
Materials and Methods
Datasets
We combined the expression files and clinical data of 697 glioma samples from TCGA (https://portal.gdc.cancer.gov/) with 207 cases of normal human cortical tissue from the GTEX (http://commonfund.nih.gov/GTEx/) database for differential analysis, as well as a training set for constructing clinical prognostic models. Two hundred and seventy-six DDR genes were labeled with the Molecular Signatures Database (MSigDB) and collected from previous study.19 We then validated the model using RNA expression and clinical data of 693 gliomas from the CGGA (http://www.cgga.org.cn) database as a validation set.
Differential, Functional and Pathway Enrichment Analyses
Differential analysis of 276 DDR-related genes was performed using the “Limma” package in R. Then, GO and KEGG enrichment analysis were performed on these obtained DDR related differential genes, using the R package ClusterProfiler. In the histogram of GO enrichment analysis, the abscissa was the number of enrichment genes, and the vertical axis was the name of enrichment pathway, which was divided into three parts: Biological process (BP), Cellular component (CC), and Molecular function (MF). The top ten significant results of each part were selected for display. The color represented the significance of the enrichment result, and the redder it was, the more significant it was. In the circle diagram, we further showed the genes corresponding to the enrichment results. In the left semicircle of the circle, we showed the enrichment of related genes. The right semicircle represented the enrichment results in different colors, corresponding to the genes on the left. We present the results of KEGG enrichment analysis using the same method.
Clinical Outcome of Distinct Molecular Subtypes Based on DDR-Related Genes
The obtained differentially expressed genes were used to stratify the glioma patients, and then survival analysis was performed for each group of patients. Consensus Clusterplus and survival package were used.
Construction and Evaluation of the DDR-Related Gene Signature
Univariate Cox and multivariate Cox survival analyses were performed on the differentially expressed genes, and K-M survival analysis was performed using the GEPIA website for each gene that has a significant prognosis after multivariate Cox screening. Then, we created an optimal risk signature with the least absolute shrinkage and selection operator (Lasso) based on “glmnet” and “survival” package. Risk score=∑(Cor*xi). Glioma patients in the training and validation sets were divided into high-risk and low-risk groups based on the median value of the DDR gene risk score. Survival analysis was performed by grouping DDR gene expression levels to evaluate the predictive efficacy of DDR gene signature for survival. In addition, receiver operating characteristics (ROC) analysis was used to assess the accuracy of the gene signature in predicting 1-, 3-, and 5-year survival rates.
Clinical Features of the DDR-Related Gene Signature
According to the high- and low-risk groups, we drew a heatmap of the correlation between the two groups and the clinicopathological parameters with “heatmap” package in R.
Construction and Evaluation of the Clinical Predicted Model
Firstly, we analyzed all possible prognostic factors using Cox regression analysis. Then, Lasso regression analysis was performed, which can increase the stability of the model by simultaneously processing multiple variables, as compared to the multi-factor Cox model, which processes multiple factors step by step. We then constructed a nomogram based on the above identified prognostic factors using the rms package in R, which was evaluated using ROC, C-index, and calibration curves. In addition, we obtained corresponding clinical information from the CGGA database for external validation. The C-index of the clinical predicted model was calculated based on CGGA data, and the calibration curve was plotted. The risk score corresponding to each clinical risk factor was obtained in the nomogram. These data were compared with the clinical predicted model constructed based on TCGA data for external validation.
Gene Set Enrichment Analysis (GSEA)
The differences of the enrichment pathways between the high-risk and low-risk groups were shown by the gene sets stemmed from the Molecular Signatures Database. Then, we determined the cutoff values of the number of permutations = 1000, and a false discovery rate (FDR) <0.25.
Real-Time PCR for Glioma Tissues
A total of 20 glioma tissue samples, including eight glioblastoma, six low-grade gliomas, and six glioma paracancerous tissues, were evaluated by RT-PCR. The use of tissue samples in this study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (No. AF-SOP-07-1.1-01).
Total RNA was extracted using RNAiso Plus reagent (Takara, No. 9108/9109). The cDNA were synthesized from ABM (Richmond, No. G592, G891, G892) and the experiments were performed according to the manufacturer’s instructions. Based on online analysis of GEPIA database (http://gepia2.cancer-pku.cn/#index), we selected 6 DDR-related genes with significant expression differences for experimental verification. The sequences of genes and internal references involved in RT-PCR experiments are shown in Table 1. The relative expression of gene determined by RT-PCR was analyzed using the 2−ΔΔCt method.
 |
Table 1 The Primer Sequences of RT-PCR |
Statistical Analysis
All statistical analyses were performed using R or R software packages, including: “limma”, “pheatmap”, “rms”, “BiocManager”, “Consensus ClusterPlus”, “survival”, “glmnet”, “survivalROC”, “forestplot”, “digest”, “GOplot”, “colorspace”, “stringi”, “ggplot2”. For all statistics in this paper, P < 0.05 was defined as statistically significant difference. In addition, PCR results were analyzed and mapped using GraphPad Prism 8 software.
Results
Differential Expression, Function and Pathway Enrichment of DDR-Related Genes
We analyzed the differential expression of 276 DDR genes, of which 75 showed differences between normal and tumor tissues (Supplementary Material 1). P < 0.05 was considered as the statistical standard (Figure 1). GO enrichment analysis showed that the 75-DDR related differential genes mainly participated in biological process involved in DNA recombination, DNA replication, double-strand break repair, telomere maintenance, telomere organization, etc. The molecular functions of the 75-DDR genes were mainly enriched in catalytic activity, acting on DNA. In addition, their cell components were mainly concentrated in replication fork, nuclear replication fork and chromosome, telomeric region, etc. (Figure 2A and B). KEGG pathway enrichment analysis suggested that these 75-DDR genes were mainly enriched in the following pathways, such as nucleotide excision repair, base excision repair and Fanconi anemia pathway, etc. (Figure 2C and D).
 |
Figure 1 A heatmap of 75 differential DDR-related genes. |
Clinical Outcomes of Distinct Molecular Subtypes Based on DDR-Related Genes
To determine whether DDR-related differential genes affect the prognosis of patients with glioma, we divided glioma patients into two groups using 75 differentially expressed genes (Figure 3A–C), and we found that there was a significant difference in prognosis between the two groups (Figure 3D).
Construction and Evaluation of DDR-Related Gene Signature
A total of 16 genes with different prognosis of glioma were screened by univariate Cox and multivariate Cox analyses, and K-M survival analysis was performed on the GEPIA website (Table 2). Using Lasso regression, fifteen genes were condensed to obtain the 12 most significant genes for prognosis, and an optimal risk signature was created. Risk score = CDC25A*0.743625+ CDC5L*(−1.14751)+ ELOA*(−0.13005)+ ERCC5*(−0.35555)+ GADD45G*(−0.20809)+ PARP4*(0.363819)* RECQL*(−0.06961)+ RFC2* 0.161676+ RPA1* 0.315634+ RRM1*(−0.34186)+ TOP3B*(−3.22014)+ WEE1* 0.650775 (Figure 4). The AUC values of the ROC curve of one year, three years and five years were 0.938, 0.851 and 0.796, respectively (Figure 5A–C). The high- and low-risk group based on the 12 genes can significantly predict the prognosis of patients with glioma (P < 0.0001) (Figure 5D).
 |
Table 2 Multivariate COX and K-M Survival Analysis |
Clinical Features of the DDR-Related Gene Signature
The DDR-related gene signature was significantly associated with patients’ clinicopathological parameters, including age (P < 0.01), grade (P < 0.001), IDH status (P < 0.001) and 1p19q codeletion status (P < 0.001) (Figure 6A). The distribution of DDR gene expression is shown in Figure 6B. Based on this DDR-related gene signature, glioma patients had significantly worse survival as the risk score increased (Figure 6C and D).
Construction and Evaluation of the Clinical Prediction Model
First, X-Tile software was used to convert the continuous variable age into a categorical variable, and the optimal cut-off value was 61 years old (Figure 7). Next, univariate Cox regression analysis was performed on clinical risk factors including age, grade, IDH status, 1p19q codeletion status, gender, radiotherapy, chemotherapy and DDR gene signature. The factors of age, grade, IDH status, 1p19q codeletion status and DDR gene signature were found to be statistically significant. On this basis, Lasso regression was used to construct a risk prediction model based on these five risk factors (Figure 8A). According to the results of Lasso regression, the risk coefficients corresponding to these five variables were age (0.064), grade (0.996), IDH status (0.150), 1p19q codeletion status (0.067) and DDR gene signature (0.239) (Figure 8B and C).
Next, we made use of these five risk factors to construct the nomogram (Figure 9A), and calculated the risk score corresponding to each risk factor in the training set and verification set respectively (Table 3). At the same time, external verification was carried out for the nomogram (Supplementary Material 2). The C-index of the training set and verification set was 0.761 (95% CI: 0.723–0.796) and 0.746 (95% CI: 0.722–0.770), respectively. The values of both were close and both were greater than 0.7, indicating that the model has been well verified. In addition, calibration curves were also drawn for the training set and verification set. It can be seen from Figure 9B and C that the calibration trends of both were close to the standard curves.
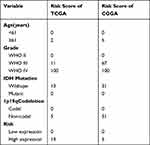 |
Table 3 Prognostic Risk Score from Nomograms |
The Analysis of the Signaling Pathways of the Different Risk Groups
GSEA was used to determine which pathways the high- and low-risk groups enriched in. The high-risk group primarily enriched in the pathways of cell cycle, DNA replication, mismatch repair, base excision repair, P53 signaling pathway, and bladder cancer. However, the low-risk group mainly enriched the following pathways, including wnt signaling pathway, ERBB signaling pathway, mTOR signaling pathway, endometrial cancer, tight junction, and hedgehog signaling pathway (Figure 10A and B; Tables 4 and 5).
 |
Table 4 The Outcome of GSEA of the High Groups |
 |
Table 5 The Outcome of GSEA of the Low Groups |
 |
Figure 10 The analysis of signaling pathways between the high- and low-groups. (A) The signaling pathways of the high groups. (B) The signaling pathways of the low groups. |
PCR Validation of DDR Gene Expression in Glioma
Total RNA from primary glioma samples was analyzed by spectrophotometry and the OD260/OD280 ratio was found to be between 1.9 and 2.0. PCR experiment results showed that six DDR genes were expressed differently between the 6 adjacent normal tissues and 14 glioma tissues. The corresponding P values of relevant genes were as follows: P(WEE1)=0.0002 (adjusted P(WEE1)= 0.0041), P(RECQL)=0.0117 (adjusted P(RECQL)= 0.0403), P(RPA1)=0.021 (adjusted P((RPA1)= 0.0460), P(RRM1)=0.0035 (adjusted P(RRM1)= 0.0403), P(PARP4)=0.0006 (adjusted P(PARP4)= 0.0099), P(ELOA)=0.0023 (adjusted P(ELOA)= 0.0296) (Figure 11). In addition, GEPIA analysis also showed significant differences in the expression of these six genes between glioma and normal tissues (Figure 12).
 |
Figure 11 qPCR verification results of the mRNA expression of six DDR-related genes. (A) WEE1, (B) RECQL, (C) RPA1, (D) RRM1, (E) PARP4, (F) ELOA. |
 |
Figure 12 Differential expression analysis of DDR-related genes using GEPIA. (A) WEE1, (B) RECQL, (C) RPA1, (D) RRM1, (E) PARP4, (F) ELOA. |
Discussion
Glioma is one of the most common cause of cancer-related deaths among brain malignancies, and it is difficult to predict its prognosis due to the phenotypic and molecular diversity. A multi-gene signature can improve the predictive ability, leading to a more robust predictive power versus a single gene. Application of prognostic model is helpful to guide clinical decisions and is essential for individualized treatment of cancer. Previous researches have shown that DNA repair systems play a vital role in cancers,20,21 thus targeting the specific genes in DDR process has potential of clinical application and targeted drugs development.22,23 In this study, for the first time, we constructed a reliable 12-DDR-related gene signature to predict the prognosis of glioma patients, which could assist the staging system to predict the clinical outcomes, thereby providing more appropriate treatment methods.
Firstly, 75 of all the 276 DDR-related genes showed differences between normal and glioma tissues. GO and KEGG enrichment analyses of 75 DDR-related DEGs showed that these genes mainly participated in DNA recombination, replication, repair, and telomere maintenance, organization, etc. Telomere can maintain chromosome stability, promote cell survival and proliferation, and prevent degenerative diseases and cancers.24,25 It acts at the ends of chromosomes to prevent them from being mistaken for chromosome breakage by DNA damage response and repair mechanisms. On the contrary, many DNA repair proteins have been found to be important for maintaining telomere structure and function. The intersection of telomere biology and DNA repair may have significant implications for human cancers. For example, telomere shortening and dysfunction may exacerbate skin pathologies.26 However, it is still unclear regarding the interacting mechanism of DNA damage and telomeres in glioma tumorigenesis. Our findings suggested the probable function and the interaction of these key genes in glioma, which is worthy to make further studies.
In addition, based on these 75 DEGs, glioma patients can be divided into two subgroups with significantly distinct clinical outcomes, which could be used to improve the stratification and individual therapeutic strategies of glioma patients. According to previous researches, the prognostic potential of DNA repair genes has been found in gastric cancer,27 colon cancer,28 hepatocellular carcinoma29 and lung adenocarcinoma,30 etc. In the present study, based on TCGA and GTEX datasets, univariate, multivariate Cox, and Lasso regression analyses, we identified a 12-DDR-related gene signature from the 75 DEGs to stratify the survival risk of glioma patients, and the overall survival of high-risk group was significantly shorter than that of low-risk group. Besides, according to the risk score assessment, patients in high- or low-risk group also had significant correlation in clinicopathological parameters, including X1p19qCodeletion, IDH_Mutation, grade and age. Furthermore, we integrated the predictive signature and clinical characteristics to construct a novel nomogram to predict overall survival. The nomogram provided favorable C-index, ROC and calibration plots, and also had a good performance in external validation cohorts of CGGA datasets, demonstrating a favorable clinical predictive ability. We further performed a GSEA analysis to explore the differences in signaling pathways for high- and low-risk groups. As a result, pathways including cell cycle, DNA replication, mismatch repair, base excision repair and P53 signaling pathway were distinctly enriched in the high-risk group. On the other hand, wnt signaling pathway, ERBB signaling pathway and MTOR signaling pathway were enriched in the low-risk group, suggesting that DNA repair genes in high- and low-risk groups participated in distinct pathways. Our model can accurately predict glioma patients’ prognostic risk based on DDR-related DEGs between high- and low-risk groups, which should be validated by in-depth analysis in the future.
We identified 12 DDR-related genes (CDC25A, CDC5L, ELOA, ERCC5, GADD45G, PARP4, RECQL, RFC2, RPA1, RRM1, TOP3B, WEE1) making up the prognostic signature, some of which were associated with glioma. Cell division cycle 25A (CDC25A), a member of the CDC25 family of phosphatases, was identified as an oncogene, and its over-expression can promote tumorigenesis.31 CDC25A over-expression activated the PI3K/AKT pathways to regulate the malignant behavior of glioma stem cells. Cell division cycle 5-like (CDC5L) protein is a cell cycle regulator of the G2/M transition and has been reported to participate in the catalytic step of pre-mRNA splicing and DNA damage repair, and knockdown of CDC5L would inhibit proliferation of glioma cells.32 GADD45 family is involved in cell cycle control, apoptosis, and repair of DNA damage.33 Cucurbitacin E can inhibit tumor growth by arresting the cell cycle at G2/M phase via GADD45G gene expression in brain malignant glioma cells.34 RecQ helicases are a ubiquitous family of DNA unwinding enzymes fulfilling similar functions in the maintenance of chromosome stability.35 RECQL is highly expressed in glioblastoma and plays an important role in tumor cell proliferation.36 Replication factor C (RFC) is a DNA-binding protein in the process of DNA repair and replication, and RFC2 is one of the five subunits of RFC.37 It is reported that RFC2 may act as an oncogene in glioblastoma progression.38 Replication protein A (RPA) is a heterotrimeric single-stranded DNA-binding protein, it is required for DNA repair pathways.39 RPA was preferentially expressed by glioblastoma stem-like cells and high RPA expression informed poor glioma patients’ survival.40 Ribonucleotide reductase catalytic subunit M1 (RRM1) plays a vital role in DNA synthesis and cell proliferation.41 RRM1 expression has been found significantly higher in glioma than in normal brain tissues, and silencing RRM1 decreased proliferation and suppressed cell cycle progression of glioma cells.42 WEE1 (WEE1 G2 checkpoint kinase) is a member of the tyrosine kinase family, it is involved in the regulation of cell cycle progression. Several studies have found that WEE1 was overexpressed in glioma and promoted glioma cells proliferation, migration, and invasion.43,44 However, the roles of some of the candidate genes in glioma have not been revealed. Nevertheless, our analysis suggested that these DDR-related genes might provide promising clinical values in glioma.
Conclusions
In conclusion, the current study proposed a novel 12 DDR-related gene signature as a practical prognostic predictor for glioma patients. A nomogram combining the signature and clinical parameters was established as an individual clinical prediction tool, which can facilitate the individualized consultation and treatment of glioma patients.
Data Sharing Statement
The following information was supplied regarding data availability: the data is available at the TCGA (https://portal.gdc.cancer.gov/), GTEX (http://commonfund.nih.gov/GTEx/) and CGGA (http://www.cgga.org.cn).
Ethics Statement
The tumor tissue used in this study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of China Medical University. This study was conducted in accordance with the Declaration of Helsinki. All patients signed informed consent forms.
Acknowledgments
The authors gratefully acknowledge financial grants from the National Natural Science Foundation of China (grant no. 31800124), the Key Project of the Natural Science Foundation of Liaoning Province (Ref No. 20180540037), and the Fundamental Computing Education Project of Association of Fundamental Computing Education in Chinese Universities (grant no. 2021-AFCEC-310).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
1. Ostrom Q, Gittleman H, Stetson L, Virk S, Barnholtz-Sloan J. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi:10.1007/978-3-319-12048-5_1
2. Jiang T, Mao Y, Ma W, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–273. doi:10.1016/j.canlet.2016.01.024
3. Ibarra L, Vilchez M, Caverzán M, Milla Sanabria L. Understanding the glioblastoma tumor biology to optimize photodynamic therapy: from molecular to cellular events. J Neurosci Res. 2021;99(4):1024–1047. doi:10.1002/jnr.24776
4. Andersen B, Miranda C, Hatzoglou V, DeAngelis L, Miller A. Leptomeningeal metastases in glioma: the Memorial Sloan Kettering Cancer Center experience. Neurology. 2019;92(21):e2483–e91. doi:10.1212/wnl.0000000000007529
5. Aldape K, Brindle K, Chesler L, et al. Challenges to curing primary brain tumours. Nat Rev. 2019;16(8):509–520. doi:10.1038/s41571-019-0177-5
6. Tolan H, Tozan-Beceren A, Sardas S, et al. DNA repair capacity of the colorectal cancer patients and the correlation between the pathological parameters. Niger J Clin Pract. 2019;22(2):194–200. doi:10.4103/njcp.njcp_251_18
7. Damia G, Broggini M. Platinum resistance in ovarian cancer: role of DNA repair. Cancers. 2019;11(1):119. doi:10.3390/cancers11010119
8. Chen X, Chen F, Ren Y, et al. IL-6 signaling contributes to radioresistance of prostate cancer through key DNA repair-associated molecules ATM, ATR, and BRCA 1/2. J Cancer Res Clin Oncol. 2019;145(6):1471–1484. doi:10.1007/s00432-019-02917-z
9. López-Camarillo C, Rincón D, Ruiz-García E, Astudillo-de la Vega H, Marchat L. DNA repair proteins as therapeutic targets in ovarian cancer. Curr Protein Pept Sci. 2019;20(4):316–323. doi:10.2174/1389203719666180914091537
10. Liu K, Jiang YJC. Polymorphisms in DNA repair gene and susceptibility to glioma: a systematic review and meta-analysis based on 33 studies with 15 SNPs in 9 genes. Neurobiology. 2017;37(2):263–274. doi:10.1007/s10571-016-0367-y
11. Liu Y, Scheurer M, El-Zein R, et al. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomark Prev. 2009;18(1):204–214. doi:10.1158/1055-9965.Epi-08-0632
12. Yosunkaya E, Kucukyuruk B, Onaran I, Gurel C, Uzan M, Kanigur-Sultuybek GJ. Glioma risk associates with polymorphisms of DNA repair genes, XRCC1 and PARP1. Br J Neurosurg. 2010;24(5):561–565. doi:10.3109/02688697.2010.489655
13. Tang L, Deng L, Bai H, et al. Reduced expression of DNA repair genes and chemosensitivity in 1p19q codeleted lower-grade gliomas. J Neurooncol. 2018;139(3):563–571. doi:10.1007/s11060-018-2915-4
14. Wang R, Ye X, Zhao X, Liu J, Zhang C. Development of a five-gene signature as a novel prognostic marker in ovarian cancer. Neoplasma. 2019;66(3):343–349. doi:10.4149/neo_2018_180705N447
15. Zheng T, Pang Z, Zhao Z. A gene signature predicts response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Biosci Rep. 2019;39(5). doi:10.1042/bsr20190414
16. Zhang D, Zheng Y, Yang S, et al. Identification of a novel glycolysis-related gene signature for predicting breast cancer survival. Front Oncol. 2020;10:596087. doi:10.3389/fonc.2020.596087
17. Sun L, Wu J, Wu Z, et al. A three-gene signature and clinical outcome in esophageal squamous cell carcinoma. Int J Cancer. 2015;136(6):E569–77. doi:10.1002/ijc.29211
18. Zuo S, Dai G, Ren X. Identification of a 6-gene signature predicting prognosis for colorectal cancer. Cancer Cell Int. 2019;19:6. doi:10.1186/s12935-018-0724-7
19. Knijnenburg T, Wang L, Zimmermann M, et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23(1):239–54.e6. doi:10.1016/j.celrep.2018.03.076
20. Mei L, Zhang J, He K, Zhang J. Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: where we stand. J Hematol Oncol. 2019;12(1):43. doi:10.1186/s13045-019-0733-6
21. Tessoulin B, Moreau-Aubry A, Descamps G, et al. Whole-exon sequencing of human myeloma cell lines shows mutations related to myeloma patients at relapse with major hits in the DNA regulation and repair pathways. J Hematol Oncol. 2018;11(1):137. doi:10.1186/s13045-018-0679-0
22. Nickoloff J, Jones D, Lee S, Williamson E, Hromas R. Drugging the cancers addicted to DNA repair. J Natl Cancer Inst. 2017;109(11). doi:10.1093/jnci/djx059
23. O’Connor M. Targeting the DNA damage response in cancer. Mol Cell. 2015;60(4):547–560. doi:10.1016/j.molcel.2015.10.040
24. Armanios M, Blackburn E. The telomere syndromes. Genetics. 2012;13(10):693–704. doi:10.1038/nrg3246
25. Duvic B, Brehélin M. Two major proteins from locust plasma are involved in coagulation and are specifically precipitated by laminarin, a beta-1,3-glucan. Insect Biochem Mol Biol. 1998;28(12):959–967. doi:10.1016/s0965-1748(98)00084-8
26. Fouquerel E, Opresko P. Convergence of the nobel fields of telomere biology and DNA repair. Photochem Photobiol. 2017;93(1):229–237. doi:10.1111/php.12672
27. Jinjia C, Xiaoyu W, Hui S, et al. The use of DNA repair genes as prognostic indicators of gastric cancer. J Cancer. 2019;10(20):4866–4875. doi:10.7150/jca.31062
28. Wang X, Tan C, Ye M, et al. Development and validation of a DNA repair gene signature for prognosis prediction in Colon Cancer. J Cancer. 2020;11(20):5918–5928. doi:10.7150/jca.46328
29. Li N, Zhao L, Guo C, Liu C, Liu Y. Identification of a novel DNA repair-related prognostic signature predicting survival of patients with hepatocellular carcinoma. Cancer Manag Res. 2019;11:7473–7484. doi:10.2147/cmar.s204864
30. Hu B, Liu D, Liu Y, Li Z. DNA repair-based gene expression signature and distinct molecular subtypes for prediction of clinical outcomes in lung adenocarcinoma. Front Med. 2020;7:615981. doi:10.3389/fmed.2020.615981
31. Li Y, Ji S, Fu L, Jiang T, Wu D, Meng F. Knockdown of cyclin-dependent kinase inhibitor 3 inhibits proliferation and invasion in human gastric cancer cells. Oncol Res. 2017;25(5):721–731. doi:10.3727/096504016x14772375848616
32. Chen W, Zhang L, Wang Y, et al. Expression of CDC5L is associated with tumor progression in gliomas. Tumour Biol. 2016;37(3):4093–4103. doi:10.1007/s13277-015-4088-5
33. Moskalev A, Smit-McBride Z, Shaposhnikov M, et al. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res Rev. 2012;11(1):51–66. doi:10.1016/j.arr.2011.09.003
34. Hsu Y, Chen M, Huang T. Inducement of mitosis delay by cucurbitacin E, a novel tetracyclic triterpene from climbing stem of Cucumis melo L., through GADD45γ in human brain malignant glioma (GBM) 8401 cells. Cell Death Dis. 2014;5:e1087. doi:10.1038/cddis.2014.22
35. Hickson I. RecQ helicases: caretakers of the genome. Cancer. 2003;3(3):169–178. doi:10.1038/nrc1012
36. Mendoza-Maldonado R, Faoro V, Bajpai S, et al. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol Cancer. 2011;10:83. doi:10.1186/1476-4598-10-83
37. Li Y, Gan S, Ren L, et al. Multifaceted regulation and functions of replication factor C family in human cancers. Am J Cancer Res. 2018;8(8):1343–1355.
38. Ho K, Kuo T, Lee Y, et al. Xanthohumol regulates miR-4749-5p-inhibited RFC2 signaling in enhancing temozolomide cytotoxicity to glioblastoma. Life Sci. 2020;254:117807. doi:10.1016/j.lfs.2020.117807
39. Ubhi T, Brown G. Exploiting DNA replication stress for cancer treatment. Cancer Res. 2019;79(8):1730–1739. doi:10.1158/0008-5472.can-18-3631
40. Pedersen H, Anne Adanma Obara E, Elbæk K, Vitting-Serup K, Hamerlik P. Replication Protein A (RPA) mediates radio-resistance of glioblastoma cancer stem-like cells. Int J Mol Sci. 2020;21(5):1588. doi:10.3390/ijms21051588
41. Kim S, Park E, Joo H, et al. RRM1 maintains centrosomal integrity via CHK1 and CDK1 signaling during replication stress. Cancer Lett. 2014;346(2):249–256. doi:10.1016/j.canlet.2013.12.031
42. Jiang K, Zhi T, Xu W, et al. MicroRNA-1468-5p inhibits glioma cell proliferation and induces cell cycle arrest by targeting RRM1. Am J Cancer Res. 2017;7(4):784–800.
43. Music D, Dahlrot R, Hermansen S, et al. Expression and prognostic value of the WEE1 kinase in gliomas. J Neurooncol. 2016;127(2):381–389. doi:10.1007/s11060-015-2050-4
44. Caretti V, Hiddingh L, Lagerweij T, et al. WEE1 kinase inhibition enhances the radiation response of diffuse intrinsic pontine gliomas. Mol Cancer Ther. 2013;12(2):141–150. doi:10.1158/1535-7163.mct-12-0735
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








