Back to Journals » Drug Design, Development and Therapy » Volume 18
A Novel DNA Aptamer Probe Recognizing Castration Resistant Prostate Cancer in vitro and in vivo Based on Cell-SELEX
Authors Zhong J , Liu D, Yang Q , Ding J, Chen X
Received 27 November 2023
Accepted for publication 9 March 2024
Published 18 March 2024 Volume 2024:18 Pages 859—870
DOI https://doi.org/10.2147/DDDT.S444988
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Manfred Ogris
Jinman Zhong,1 Duoduo Liu,1 Quanxin Yang,1 Jianke Ding,2 Xin Chen1
1Department of Radiology, The Second Affiliated Hospital, Xi’ an Jiaotong University, Xi’an, Shaanxi Province, 710004, People’s Republic of China; 2Department of Plastic and Reconstructive Surgery, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi Province, 710032, People’s Republic of China
Correspondence: Xin Chen, Department of Radiology, The Second Affiliated Hospital, Xi’ an Jiaotong University, 157 Xiwu Road, Xi’an, Shaanxi Province, 710004, People’s Republic of China, Email [email protected] Jianke Ding, Department of Plastic and Reconstructive Surgery, Xijing Hospital, Fourth Military Medical University, 127 Changle West Road, Xi’an, Shaanxi Province, 710032, People’s Republic of China, Email [email protected]
Background: Early recognition of castration-resistant state is of significance for timely adjustment of treatment regimens and improvement of prognosis.
Purpose: This study aims to screen new aptamers CRda8 and CRda21 which recognize castration resistant prostate cancer (CRPC) cells with high affinity and specificity by SELEX technology.
Methods: The enrichment of specific aptamer candidates was monitored by flow cytometric analysis. The affinity and specificity of aptamer candidates were evaluated by flow cytometry and immunofluorescence assay. MR imaging of CRda21-conjugated polyethylene glycol (PEG)-Fe3O4 nanoparticles to CRPC was further explored in vivo.
Results: Both aptamers showed high specificity to target cells with dissociation constants in the nanomolar range, and did not recognize other tested cells. The staining of clinical tissue sections with fluorescent dye labeled aptamers showed that sections from CRPC exhibited stronger fluorescence while sections from benign prostatic hyperplasia and androgen dependent prostate cancer did not exhibit notable fluorescence. In vivo MRI demonstrated that CRda21-conjugated PEG-Fe3O4 had good affinity to CRPC and produced strong T2WI signal intensity reduction distinguished from peritumoral tissue.
Conclusion: The high affinity and specificity of CRda8 and CRda21 make the aptamer hold potential for early recognition of castration-resistant state and diagnosis of CRPC at the cellular level.
Keywords: aptamer, Cell-SELEX, MRI, castration resistant prostate cancer, androgen dependent prostate cancer
Introduction
Prostate cancer is one of the most common malignancies in males worldwide. According to the annual reports from the American Cancer Society, prostate cancer incidence increased by 3% annually from 2014 through 2019 after two decades of decline.1 There are no initial or early symptoms in most cases. Diagnosis is primarily based on prostate-specific antigen (PSA) testing and transrectal ultrasound-guided prostate tissue biopsies. Advances in screening and diagnosis have allowed detection and treatment of the disease in early stage, especially in western countries. In Asian countries, lots of patients are often diagnosed with late-stage diseases on the first visit, mostly because of relatively backward economic development and imperfect cancer screening system.2
Androgen deprivation therapy (ADT) is typically used in advanced prostate cancer as a means of slowing tumor progression and alleviating symptoms. Initial responses to castration therapy are quite favorable, with a significant clinical regression and rapid biochemical response.3,4 However, these patients may eventually progress to an aggressive and metastatic state refractory to androgen deprivation (referred to as castration resistant prostate cancer, CRPC), which carries a worse prognosis and increases the mortality burden of the population.5 Recent evidences suggest that ADT may cause cadherin switching from E-cadherin to N-cadherin, which is a key process of epithelial-mesenchymal transition, leading to a progressive development prior to morphology in prostate cancer migration and invasion.6 According to the guidelines on the treatment of relapsing, metastatic, and CRPC of the European Association of Urology,7 CRPC is defined as castrate serum testosterone < 50 ng/dl plus one of the following types of progression: (1) Biochemical progression: three consecutive rises in PSA 1 week apart, resulting in two 50% increases over the nadir, and PSA > 2 ng/mL; (2) Radiologic progression: the appearance of new lesions: either two or more new bone lesions on bone scan or a soft tissue lesion using the Response Criteria in Solid Tumors.8 However, the transition of androgen dependent prostate cancer (ADPC) cells into CRPC cells is prior to biochemical or radiologic changes, thus morphologic and symptomatic progressions are not sufficient for diagnosing CRPC in early.6,9 Furthermore, the present imaging systems, including ultrasound, computed tomography (CT) and magnetic resonance imaging (MRI) are used for routine diagnosis of prostate cancer, which is almost impossible for identification of different state of prostate cancer progression. On that basis, early recognition of castration-resistant state and diagnosis of CRPC at the cellular level is of significance for timely adjustment of treatment regimens and improvement of prognosis.
Aptamers are single- or double-stranded DNA or RNA oligonucleotides which have attracted much attention as a new class of molecular probes for tumor diagnosis and treatment in molecular medicine.10,11 Compared with antibodies, aptamers have excellent chemical properties such as easy synthesis, controllable modifications, high stability, low immunogenicity and fast tissue penetration.12–17 Aptamers are derived from systematic evolution of ligands by exponential enrichment (SELEX) technology. Cell-SELEX is a modification of the traditional SELEX procedure using whole living cells as target, which generates cell-specific aptamers by employing the differences at the molecular level between any two cell lines.18 Aptamers screened by Cell-SELEX recognize the target cells without prior knowing the exact membrane proteins, which is helpful to generate cell-specific aptamers targeting CRPC cells.
In this study, we selected a DNA aptamer by Cell-SELEX, which could recognize C4-2 cells (a CRPC cell line) and distinguish CRPC samples from ADPC samples with high affinity and specificity in vitro and in vivo.
Materials and Methods
Cell Lines and Cell Culture
Human prostate cancer C4-2, PC-3, DU145 and LNCap cell lines, human prostate stromal myofibroblastic immortalized cell lines WPMY-1, and human hepatic carcinoma SMMC7721 cell lines were purchased from Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China). DU145, LNCap and WPMY-1 cells were cultured in the basic medium of Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA); C4-2, PC-3 and SMMC7721 cells were cultured in the medium of RPMI-1640 (Gibco, USA) with 10% FBS. All cell lines were maintained at 37°C under a mixture of 95% air and 5% CO2.
Selection of Aptamers for CRPC Cells
Selection of aptamers for CRPC cells was accomplished by Cell-SELEX. The initial single-stranded DNA (ssDNA) library (5′-TGCGGCAGTTGAAGCAAGGC-40N-ACGGCAGCACCAGAGAACCA-3′) was synthesized by Sangon Biotech Co. Ltd. (Shanghai, China) and purified by HPLC. The forward primer sequence was 5′-FAM-TGCGGCAGTTGAAGCAAGGC-3′, and the reverse primer sequence was 5′-Biotin-TGGTTCTCTGGTGCTGCCGT-3′. Cells were incubated with the initial library for 1 hour at 4°C in selection buffer (0.1 M Hepes, 0.75 M NaCl, 0.025 M KCl, 0.01 M CaCl2, 0.01 M MgCl2·6H2O in double distilled water, pH 7.4). In the first round, C4-2 cells were detached with a cell scraper and binding ssDNAs were eluted with PBS for 5 minutes. After centrifugation at 12,000 g for 5 minutes at 4°C, eluted nucleic acids were amplified by PCR under the following conditions: 95°C for 30 minutes, 10–15 cycles of 30 seconds at 95°C, 30 seconds annealing at 60°C, and 1 minute extension at 72°C, followed by 72°C for 5 minutes. The PCR product was precipitated with sodium acetate and cooled isopropanol at −20°C for 3 h, and then centrifuged at 12,000 g for 10 minutes at 4°C, after which the supernatant was removed and the DNA pellet was washed with 1 mL 75% cooled ethanol and then resuspended in 200 µL of Buffer I (10 mM Tris-HCl, 1 mM EDTA, 1 M NaCl, 0.01–0.1% Tween-20 in double distilled water). The DNA was incubated with streptavidin-coated sepharose beads for 30 minutes at room temperature, then denatured by incubation with 0.15 M NaOH for 20 minutes. The selected ssDNA was lyophilized and resuspended in binding buffer for the subsequent selection procedure. The cell-SELEX selection process is similar to our previous study in which we successfully selected the EpCAM-specific DNA aptamers recognizing prostate cancer cells.19
Beginning with the sixth round, the ssDNA pool was incubated with LNCap cells (negative control cells) and the unbound oligonucleotides were collected and incubated with C4-2 cells for positive selection. A total of 14 rounds of selection were done. The clone and sequencing were performed by Sangon Biotech Co. Ltd.
Flow Cytometry for Identification of Target Aptamers
For enrichment analysis, 2×105 C4-2 and LNCap cells were respectively incubated with each round of the selected sequences at 4°C for 30 minutes. The selected sequences and the initial ssDNA library were FAM-labeled. The suspended cells were rinsed with washing buffer three times for 5 minutes to remove unconjugated ligands and resuspended in 200 µL of binding buffer. The fluorescence signal was analyzed with a FACSCalibur flow cytometer (BD Biosciences, USA).
Effect of Temperature on Aptamers Binding Ability
To investigate whether incubation temperature would affect the binding capacity of aptamers, 1 × 105/mL C4-2 and LNCap cells were respectively seeded in a 20 mm glass bottom dish and maintained overnight. The cells were fixed in 4% paraformaldehyde in PBS for 15 minutes and washed three times with PBS, following which the cells were blocked with 5% BSA in PBS at room temperature for 1 hour and stained with 200 pmol of FAM-labeled aptamers at 4°C or 37°C for 30 minutes. Nuclei were counterstained with 10 µg/mL DAPI (Invitrogen, Carlsbad, CA, USA). After washing, the stained cells were imaged by confocal laser scanning microscope (Olympus, Japan).
To measure the equilibrium dissociation constant (Kd) of aptamers, C4-2 cells were incubated with a series of concentration gradients of FAM-labeled aptamers (0, 0.5, 5, 10, 15, 20, 25, 30, 35, 50, 75, 100, 125, 150, 175, 200, 250, 300 nM) at 4°C or at 37 °C for 1 hour. Cells were rinsed with washing buffer three times for 5 minutes and resuspended in 200 µL of binding buffer for flow cytometric analysis. The average fluorescence intensity at each concentration was recorded. The Kds of aptamers were determined by fitting the dependence of fluorescence intensity on aptamer concentration with the equation Y=Bmax × X/ (Kd+X), where Y is fluorescence intensity of cells at each concentration, X is concentration of the aptamer and Bmax is maximum binding potential. The experiment was performed three times independently.
Characterization of Aptamer Affinity and Specificity to Target Cells
The binding specificity of aptamer CRda8 and CRda21 to target cell line and other cell lines were evaluated by flow cytometry and confocal microscopy imaging. 2×105 C4-2, PC-3, DU145, LNCap, WPMY-1 and SMMC7721 cells (n = [3 × 6 +1] × 6×3) were respectively incubated with 200 pmol FAM-labeled aptamers at 4°C for 30 minutes. The initial ssDNA library was used as control. The cell suspension was centrifuged and washed three times for 5 minutes and resuspended in 200 µL of binding buffer. Samples were evaluated by using a FACSCalibur flow cytometer (BD Biosciences, USA). Confocal microscopy imaging of cells was also performed to investigate the affinity of aptamers, which was similar with the process as described above (2.4 Effect of temperature on aptamers binding ability): 1 × 105/mL C4-2, PC-3, DU145, LNCap, WPMY-1 and SMMC7721 cells were respectively seeded in a 20 mm glass bottom dish and maintained overnight (n = 3×8 × 6×3). The cells were fixed in 4% paraformaldehyde in PBS for 15 minutes. After washing, cells were blocked with 5% BSA in PBS at room temperature for 1 hour and stained with 200 pmol of FAM-labeled aptamers at 4°C for 30 minutes. Nuclei were counterstained with 10 µg/mL DAPI (Invitrogen, Carlsbad, CA, USA). After washing, the stained cells were imaged by confocal laser scanning microscope (Olympus, Japan). All of the experiments for the binding assay were repeated in triplicate.
Imaging of Clinical Pathological Human Prostate Tissues with Aptamers
The experiment was granted by the institutional review board of Xi’ an Jiaotong University (Xi’an, China) and informed consent has been provided by the patients. Tissues specimens of benign prostate hyperplasia (BPH), ADPC and CRPC were provided by the Department of Pathology of the Second affiliated Hospital, Xi’ an Jiaotong University. BPH specimens were obtained by transurethral resection of the prostate. ADPC specimens were obtained from patients with advanced prostate cancer who were sensitive to ADT. CRPC specimens were obtained from the patients with prostate cancer who were resistant to hormone therapy over time. Tissues specimens were fixed in 4% paraformaldehyde for 24 hour and embedded in paraffin. Tissue sections of 4 µm were cut and deparaffinated in xylene twice. Use the gradient ethanol (100%, 95%, 85% and 70%) to rehydrate the sections and rinse the slides in deionized water. The slices were stained by hematoxylin and eosin (HE) to observe the structure of the three types of tissues. For immunofluorescence staining, sections were submitted to an antigen retrieval step at 96°C for 20 minutes in citrate buffer at PH 6.0 and blocked with 5% BSA for 1 hour. 2 nM FAM-labeled aptamers were applied overnight at 4°C in a humid chamber. Nuclei were counterstained with 10 µg/mL DAPI (Invitrogen, Carlsbad, CA, USA). The stained sections (n = 2×6 × 3×3) were imaged by confocal laser scanning microscope (Olympus, Japan). The experiments were repeated three times independently.
Synthesis of Aptamer-Conjugated Magnetic Nanoparticles and in vivo MR Imaging
Biotin-labeled aptamers were reacted with streptavidin-coated polyethylene glycol (PEG)-Fe3O4 nanoparticles (Nanoeast Biological Technology Co., Ltd., Nanjing, China) for 1 hour on a shaker to synthetize composite aptamer-magnetic nanoparticles CRda21-SPION. The coupled nanoparticles were magnetically separated from free aptamers, and then rinsed with washing buffer three times and resuspended in PBS. PEG-Fe3O4 nanoparticles coupled with initial library were synthesized in the same way and used as a control.
Animal studies were conducted in accordance with the Animal Care and Use Committee of Xi’ an Jiaotong University and followed the guidelines of “Laboratory Animal-General requirements for animal experiments” (GB/T, 35823–2018, China). The Animal Care and Use Committee of Xi’ an Jiaotong University has approved the animal experiments conducted in the study. Male nude mice (Bab/ c, n = 32), aged 5–6 weeks, were used to establish the prostate tumor-bearing model with subcutaneous injection of 5×106 C4-2 cells (CRPC tumor-bearing mice) or 5×106 LNCap cells (ADPC tumor-bearing mice). Experiment was performed when tumors reached approximately 0.8–1.0 cm in diameter. CRPC tumor-bearing mice and ADPC tumor-bearing mice were respectively randomly divided into experimental group (n = 8, administered 5 mg/kg CRda21-SPION by tail vein injection) and control group (n = 8, administered 5 mg/kg PEG-Fe3O4 coupled with library by tail vein injection). The mice were anesthetized with an intraperitoneal injection of 1% sodium pentobarbital (0.5 mL/100 g) and then placed in the supine position and examined on a 3.0T MRI scanner (Siemens Magnetom Tim Trio MRI) with a small-animal coil before the injection of the probes. Subsequently, the mice respectively underwent MRI examinations at 30 minutes, 1 hour or 2 hours post-injection under the same conditions. The mice were taken out from the MRI scanner after each scan, temporarily placed in the cage and kept warm. T2-weighted imaging (T2WI) were acquired with a repetition time (TR) of 7400 ms, an echo time (TE) of 66 ms, a field-of-view (FOV) of 200 mm, slice thickness of 2.0 mm, flip angle of 150°. After MRI examination, the data were sent to a dedicated MRI image processing workstation (Leonardo; Siemens) for further analysis. Kruskal–Wallis H-test followed by Nemenyi test were used to compare in vivo T2WI signal intensity measurements in tumors at pre-injection and three post-injection time intervals. A P value less than 0.05 denoted a significant difference. All mice were euthanized by cervical dislocation after MRI examination.
Results
Selection of Aptamer Against CRPC C4-2 Cells
To screen aptamers specifically targeting CRPC, we selected CRPC cell line C4-2 to be the target cells and mediated negative selection by ADPC cell line LNCap. The process of Cell-SELEX for aptamer selection is shown in Figure 1A. For monitoring the degree of enrichment, we analyzed the ssDNA libraries derived from the fourth, eighth, eleventh and fourteenth rounds of selection by flow cytometry. As shown in Figure 1B, when the number of turns increased, the fluorescence intensity of C4-2 cells gradually increased. In contrast, the LNCap cells exhibited only weak fluorescence strength, similar to the control group (Figure 1C). These results demonstrated that the oligonucleotide products of the fourteenth round had the highest ability to bind to C4-2 cells but not to LNCap cells.
Identification of DNA Aptamers Against Target Cells
A total of 34 clones were obtained after alignment. By analyzing the predicted secondary structure of the sequences, 5 sequences were truncated and synthesized for binding assay. Flow cytometry assay showed that two sequences CRda8 and CRda21 (Table 1) exhibited the strongest affinity to C4-2 cells (Figure 2; n = [1 + 6×6] × 3). Therefore, the two sequences were selected and chemically synthesized for further characterization. The predicted secondary structure of CRda8 and CRda21 were showed in Figure 3.
 |
Table 1 Sequences of Aptamer CRda8 and CRda21 |
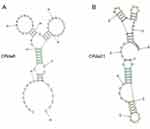 |
Figure 3 Predicted secondary structures of aptamer candidates CRda8 (A) and CRda21 (B). |
Affinity of Aptamer CRda8 and CRda21 at Different Temperatures
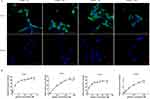
The target recognition performance of aptamer CRda8 and CRda21 after temperature shift were respectively examined. As showed in Figure 4A, the fluorescence signals of FAM-conjugated CRda8 and CRda21 were respectively clustered on the surfaces of C4-2 cells at both 4 °C and 37 °C, but not on LNCap cells. In addition, the Kds of CRda8 binding to C4-2 cells were 4.9 ± 1.6 nM at 4 °C and 101.3 ± 28.3 nM at 37 °C; the Kds of CRda21 binding to C4-2 cells were 33.26 ± 4.1 nM at 4 °C and 62.7 ± 11.7 nM at 37 °C (Figure 4B). The experiment indicated that aptamer CRda8 and CRda21 could specifically recognize C4-2 cells with high affinity and an increase in temperature has little effect on the cell recognition of the aptamers. In addition, compared with CRda8, aptamer CRda21 had higher affinity to target cells at the temperature of the human body.
Affinity and Specificity of Aptamers to Target Cells
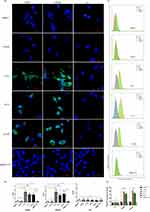
As for the cell-binding specificity of CRda8 and CRda21, WPMY-1, LNCap, C4-2, PC-3, DU145 and SMMC7721 cells were respectively incubated with FAM-labeled aptamers at 4°C for 30 minutes and analyzed by flow cytometry. As showed in Figure 5A and B, aptamer CRda8 and CRda21 showed no binding affinity to WPMY-1, LNCap and SMMC7721 cells, but showed high binding affinity to C4-2, PC-3 and DU145 cells which all belonged to human CRPC cell lines. The results from confocal microscopy imaging (Figure 5C and D) were similar to flow cytometry results. Fluorescence signals of FAM-conjugated CRda8 and CRda21 were respectively observed largely on the surfaces of C4-2, PC-3 and DU145 cells, but not on WPMY-1, LNCap and SMMC7721 cells.
Imaging of Clinical Pathological Human Prostate Tissues with Aptamers
To verify that the aptamers had the ability to recognize human CRPC tissues, confocal laser scanning microscope was used to image the three types of human prostate tissues with CRda8 and CRda21. As showed in Figure 6, CRPC tissues showed strong fluorescence signal, while BPH and ADPC tissues showed weak fluorescence signal. The results indicated that CRda8 and CRda21 had strong binding and recognition ability for human CRPC tissues.
In vivo MR Imaging of CRda21
To investigate whether the aptamers retained their recognition ability in vivo, subcutaneous C4-2 CRPC xenograft tumor model and LNCap ADPC xenograft tumor model were respectively established. As illustrated in Figure 7, the signal intensity in the tumor site significantly decreased at 30 minutes after injecting CRda21-SPION into CRPC tumor-bearing mice, and continued to decrease at 1 hour post-injection. The T2WI signal intensity changes of tumors after injection of CRda21-SPION were statistically significant when compared to the baseline pre-injection T2WI signal intensity levels of tumor (P < 0.05 or P < 0.01). The T2WI signal intensity of tumors gradually increased 2 hours later. Accumulation of CRda21-SPION was greatest in tumors with relatively little in normal tissue distribution. There were no statistically significant T2WI signal intensity changes after injection of ssDNA-SPION (All P > 0.05). In addition, the signal intensity in the tumor site slightly decreased at 30 minutes after injecting CRda21-SPION into ADPC tumor-bearing mice, but no statistical difference was found between the signal intensity at any two time points (All P > 0.05). Although the T2WI signal intensity of CRPC tumor and ADPC tumor both decreased at 30 minutes post-injection, the signal intensity of the former was significantly lower than that of the latter (P < 0.05). The results illustrated that aptamer CRda21 possessed in vivo CRPC targeting ability, providing a potential molecular probe for early diagnosis of CRPC.
Discussion
Castration resistance is defined as the progression of disease in a castration environment, and it precedes hormone resistance, which is defined as the progression of disease despite whichever hormonal manipulation is added to castration.20 Previous studies showed that 10–20% of patients with prostate cancer metastasis develop CRPC within 5 years of follow-up. Furthermore, patients with non-metastatic CRPC are at higher risk of progression.21 CRPC is considered as the terminal stage of prostate cancer. Treatment of CRPC is usually palliative, and disease evolution is often associated with significant morbidity. On that basis, early recognition of CRPC at the cellular level and the proper selection of treatment is important.
In the study, we first successfully screened CRPC-specific DNA aptamers via Cell-SELEX, which were identified to have high affinity and specificity for CRPC cells and tissues in vitro and in vivo. The scheme 1 showed the schematic representation of the research including the process of aptamer selection, aptamer-magnetic nanoparticles conjugation, probe imaging in vitro and in vivo. Our findings suggest that the aptamers have the potential for noninvasive diagnosis of CRPC in early.
 |
Scheme 1 Schematic representation of the process for the research including the process of aptamer selection, aptamer-magnetic nanoparticles conjugation, probe imaging in vitro and in vivo. |
The aptamers selected in our study were DNA aptamers. At first, RNA aptamers were preferred over DNA aptamers for their higher affinity and diversity of 3D structures. However, researchers found that RNA aptamers were easily degraded by RNase, which increased the difficulty of its screening and application. It has been gradually realized that DNA aptamers were much more stable in structures, thus were widely screened and used in practical application.22
As with antibodies, aptamers bind to the target by folding into a specific three-dimensional conformation that is dictated largely by the nucleic acid sequence.23,24 The major factor that affects affinity and dissociation constant of aptamers is temperature under the same pressure.25 Temperature changes may influence the conformation of oligonucleotide molecules and a low temperature of 4°C is often used in the Cell-SELEX screening process to inhibit endocytosis. In our study, the Kd of CRda8 and CRda21 binding to C4-2 cells were calculated as 4.9 ± 1.6 nM and 33.26 ± 4.1 nM at 4 °C, respectively. When the temperature was raised to 37°C, the Kd of CRda8 and CRda21 binding to C4-2 cells were calculated as 101.3 ± 28.3 nM and 62.7 ± 11.7 nM at 37 °C. The Kds of CRda8 and CRda21 binding to C4-2 cells are both in the nanomolar range. Although the Kd of the aptamers at 37 °C did increase compared with that at 4 °C, vivid green fluorescence of FAM-conjugated CRda8 and CRda21 were clustered on the surfaces of C4-2 cells at both 4 °C and 37 °C as illustrated in Figure 4, which demonstrates that temperature shift within the expected range does not significantly affect the affinity of CRda8 and CRda21. DNA aptamer CRda8 and CRda21 show stability after temperature shift. Considering the range of Kd variation of CRda8 is larger than that of CRda21, and the Kd value of CRda21 is lower than that of CRda8 at the temperature of the human body, we chose CRda21 for further in vivo investigation.
MRI incorporating anatomical and functional imaging has been validated as a means of detecting and characterising prostate cancer and can aid in risk stratification and treatment selection.26,27 Like many other tumors, prostate cancer shows earlier and more pronounced enhancement than surrounding normal tissues on contrast-enhanced MRI.28 Superparamagnetic iron oxide nanoparticles are known as a MRI negative contrast agent which decrease the MR signal intensity by dephasing the transverse magnetization and reducing the value of transverse relaxation time T2.29,30 In our study, polyethylene glycol (PEG)-Fe3O4, a type of superparamagnetic iron oxide nanoparticles, were used to reacted with aptamer CRda21 via biotin-streptavidin-system to synthetize composite aptamer-magnetic nanoparticles CRda21-SPION. The spin-spin relaxation time (T2) and the R2 (1/T2) of PEG-Fe3O4 nanoparticles were measured and the T2 relaxivity value of PEG-Fe3O4 was calculated as 167.2 mM−1 S−1 (Figure S1).
To further investigate whether the aptamer retained its recognition ability in vivo, subcutaneous CRPC models and ADPC models were established, and the tumor-bearing mice were respectively administrated with CRda21-SPION via tail vein. As showed in Figure 7, the tumor showed significantly decreased T2 signal intensity compared to pre-injection levels in CRPC tumor-bearing mice. The evidence indicated that CRda21 possessed in vivo CRPC targeting ability, providing a potential molecular probe for early diagnosis of CRPC. Furthermore, the toxicity profile of CRda21-SPION was studied in vitro and the data demonstrated that CRda21-SPION has effects on cells. As showed in Figure S2, the viability of C4-2 and LNCap cells treated with CRda21-SPION decreased compared with the control group (cells without treatment), but there was no significant difference between the two groups (P = 0.0609).
The study possesses limitations. First, the number of proper aptamers selected via Cell-SELEX in the study is small; only two aptamer candidates showed affinity and specificity to CRPC cells and tissues finally. In addition, the possible interaction mechanisms of aptamers over prostate cancer cells have not been investigated yet, which needs further research.
Conclusions
In summary, the DNA aptamers against CRPC have been generated by Cell-SELEX through 14 rounds of evolved enrichment. The study shows the aptamers specifically recognize target cells and tissues with high affinity. In addition, by in vivo MR imaging assay, aptamer CRda21 could accumulate at the CRPC tumor site and significantly decrease the T2WI signal intensity of the tumor, allowing clear visualization of the tumor location in vivo. Our study first selects the aptamers against CRPC, which has the potential for noninvasive diagnosis of CRPC in early at the cellular level.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant number: 82102347) and the Shanxi Provincial key research and development project (Grant number: 2024SF-YBXM-417).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi:10.3322/caac.21763
2. Hassanipour S, Delam H, Arab-Zozani M, et al. Survival rate of prostate cancer in Asian countries: a systematic review and meta-analysis. Ann Glob Health. 2020;86(1):2. doi:10.5334/aogh.2607
3. Desai K, McManus JM, Sharifi N. Hormonal therapy for prostate cancer. Endocr Rev. 2021;42(3):354–373. doi:10.1210/endrev/bnab002
4. Schaeffer E, Srinivas S, Antonarakis ES, et al. NCCN guidelines insights: prostate cancer, Version 1.2021. J Natl Compr Canc Netw. 2021;19(2):134–143. doi:10.6004/jnccn.2021.0008
5. Achard V, Putora PM, Omlin A, Zilli T, Fischer S. Metastatic prostate cancer: treatment options. Oncology. 2022;100(1):48–59. doi:10.1159/000519861
6. Varisli L, Tolan V, Cen JH, Vlahopoulos S, Cen O. Dissecting the effects of androgen deprivation therapy on cadherin switching in advanced prostate cancer: a molecular perspective. Oncol Res. 2022;30(3):137–155. doi:10.32604/or.2022.026074
7. Cornford P, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur Urol. 2017;71(4):630–642. doi:10.1016/j.eururo.2016.08.002
8. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi:10.1016/j.ejca.2008.10.026
9. Cheng Q, Butler W, Zhou Y, et al. Pre-existing castration-resistant prostate cancer-like cells in primary prostate cancer promote resistance to hormonal therapy. Eur Urol. 2022;81(5):446–455. doi:10.1016/j.eururo.2021.12.039
10. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi:10.1126/science.2200121
11. Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv. 2015;33(6 Pt 2):1141–1161. doi:10.1016/j.biotechadv.2015.02.008
12. Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48(15):2672–2689. doi:10.1002/anie.200804643
13. Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2(1):241–264. doi:10.1146/annurev.anchem.1.031207.112851
14. Xing H, Wong NY, Xiang Y, Lu Y. DNA aptamer functionalized nanomaterials for intracellular analysis, cancer cell imaging and drug delivery. Curr Opin Chem Biol. 2012;16(3–4):429–435. doi:10.1016/j.cbpa.2012.03.016
15. Song KM, Lee S, Ban C. Aptamers and their biological applications. Sensors. 2012;12(1):612–631. doi:10.3390/s120100612
16. Iliuk AB, Hu L, Tao WA. Aptamer in bioanalytical applications. Anal Chem. 2011;83(12):4440–4452. doi:10.1021/ac201057w
17. Yuan B, Zhou Y, Guo Q, et al. A signal-on split aptasensor for highly sensitive and specific detection of tumor cells based on FRET. Chem Commun. 2016;52(8):1590–1593. doi:10.1039/C5CC08060F
18. Tan Y, Guo Q, Xie Q, et al. Single-walled carbon nanotubes (SWCNTs)-assisted cell-systematic evolution of ligands by exponential enrichment (cell-SELEX) for improving screening efficiency. Anal Chem. 2014;86(19):9466–9472. doi:10.1021/ac502166b
19. Zhong J, Ding J, Deng L, et al. Selection of DNA aptamers recognizing EpCAM-positive prostate cancer by cell-SELEX for in vitro and in vivo MR Imaging. Drug Des Devel Ther. 2021;15:3985–3996. doi:10.2147/DDDT.S322854
20. Morote J, Aguilar A, Planas J, Trilla E. Definition of castrate resistant prostate cancer: new insights. Biomedicines. 2022;10(3):689. doi:10.3390/biomedicines10030689
21. Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract. 2011;65(11):1180–1192. doi:10.1111/j.1742-1241.2011.02799.x
22. Wang T, Chen C, Larcher LM, Barrero RA, Veedu RN. Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol Adv. 2019;37(1):28–50. doi:10.1016/j.biotechadv.2018.11.001
23. Guan B, Zhang X. Aptamers as versatile ligands for biomedical and pharmaceutical applications. Int J Nanomed. 2020;15:1059–1071. doi:10.2147/IJN.S237544
24. Kumar KP, Hussain B, Yuce M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics. 2020;12(7):646.
25. Sakamoto T, Ennifar E, Nakamura Y. Thermodynamic study of aptamers binding to their target proteins. Biochimie. 2018;145:91–97. doi:10.1016/j.biochi.2017.10.010
26. Wei X, Xu J, Zhong S, et al. Diagnostic value of combining PI-RADS v2.1 with PSAD in clinically significant prostate cancer. Abdom Radiol. 2022;47(10):3574–3582. doi:10.1007/s00261-022-03592-4
27. Fernandes MC, Yildirim O, Woo S, Vargas HA, Hricak H. The role of MRI in prostate cancer: current and future directions. MAGMA. 2022;35(4):503–521. doi:10.1007/s10334-022-01006-6
28. Verma S, Turkbey B, Muradyan N, et al. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR Am J Roentgenol. 2012;198(6):1277–1288. doi:10.2214/AJR.12.8510
29. Chelluri LK, Mohanram Y, Jain R, et al. Effect of engineered superparamagnetic iron oxide nanoparticles in targeted cardiac precursor cell delivery by MRI. Biochem Biophys Res Commun. 2021;541:15–21. doi:10.1016/j.bbrc.2021.01.005
30. Iacobazzi RM, Vischio F, Arduino I, et al. Magnetic implants in vivo guiding sorafenib liver delivery by superparamagnetic solid lipid nanoparticles. J Colloid Interface Sci. 2022;608(Pt 1):239–254. doi:10.1016/j.jcis.2021.09.174
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.