Back to Journals » Infection and Drug Resistance » Volume 15
A New Variant of Mutational and Polymorphic Signatures in the ERG11 Gene of Fluconazole-Resistant Candida albicans
Authors Odiba AS , Durojaye OA, Ezeonu IM, Mgbeahuruike AC, Nwanguma BC
Received 16 February 2022
Accepted for publication 3 June 2022
Published 17 June 2022 Volume 2022:15 Pages 3111—3133
DOI https://doi.org/10.2147/IDR.S360973
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Arome Solomon Odiba,1,2 Olanrewaju Ayodeji Durojaye,3– 5 Ifeoma Maureen Ezeonu,6 Anthony Christian Mgbeahuruike,7 Bennett Chima Nwanguma1,2
1Department of Biochemistry, Faculty of Biological Sciences, University of Nigeria, Nsukka, Enugu State, 410001, Nigeria; 2Department of Molecular Genetics and Biotechnology, University of Nigeria, Nsukka, Enugu State, 410001, Nigeria; 3Department of Chemical Sciences, Coal City University, Emene, Enugu State, Nigeria; 4Department of Molecular and Cell Biology, University of Science and Technology of China, Hefei, Anhui, 230026, People’s Republic of China; 5MOE Key Laboratory of Membraneless Organelle and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, University of Science and Technology of China, Hefei, Anhui, People’s Republic of China; 6Department of Microbiology, Faculty of Biological Sciences, University of Nigeria, Nsukka, Enugu State, 410001, Nigeria; 7Department of Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, University of Nigeria, Nsukka, Enugu State, 410001, Nigeria
Correspondence: Bennett Chima Nwanguma; Anthony Christian Mgbeahuruike, Department of Biochemistry, Faculty of Biological Sciences, University of Nigeria, Enugu State, Nsukka, 410001, Nigeria, Tel +234 8063655062, Email [email protected]; [email protected]
Background: Resistance to antifungal drugs for treating Candida infections remains a major concern globally despite the range of medications available. Most of these drugs target key proteins essential to the life cycle of the organism. An enzyme essential for fungal cell membrane integrity, lanosterol 14–α demethylase (CYP51), is encoded by the ERG11 gene in Candida species. This enzyme is the target of azole–based drugs. The organism has, however, devised molecular adaptations to evade the activity of these drugs.
Materials and Methods: Classical methods were employed to characterize clinical isolates sampled from women and dogs of reproductive age. For fluconazole efficacy studies, CLSI guidelines on drug susceptibility testing were used. To understand the susceptibility pattern, various molecular and structural analytic approaches, including sequencing, in silico site-directed mutagenesis, and protein-ligand profiling, were applied to the ERG11 gene and CYP51 protein sequences. Several platforms, comprising Clustal Omega, Pymol plugin manager, Pymol molecular visualizer, Chimera–curated Dynameomics rotamer library, protein–ligand interaction profiler, Charmm36 force field, GROMACS, Geneious, and Mega7, were employed for this analysis.
Results: The following Candida species distribution was obtained: 37.84% C. albicans, 8.12% C. glabrata, 10.81% C. krusei, 5.41% C. tropicalis, and 37.84% of other unidentified Candida species. Two codons in the nucleotide sequence of the wild-type (CTC and CCA) coding for LEU– 370 and PRO– 375, respectively, were mutated to L370S and P375H in the resistant strain. The mutation stabilized the protein at the expense of the heme moiety. We found that the susceptible isolate from dogs (Can–iso– 029/dog) is closely related to the most resistant isolate from humans.
Conclusion: Taken together, our results showed new mutations in the heme-binding pocket of caCYP51 that explain the resistance to fluconazole exhibited by the Candida isolates. So far, the L370S and P375H resistance-linked mutations have not been previously reported.
Keywords: Candida infection, CYP51, drug resistance, ERG11, fluconazole, mutation
Plain Language Summary
Most vaginal infections are very uncomfortable, with symptoms that include itching, pain during sexual intercourse, and soreness in the vagina. Vulvovaginal candidiasis is one such infection in which a serious condition could lead to vaginal cracks. This infection is caused by different species of the microorganism known as Candida, of which the most common is Candida albicans. Several drugs, including fluconazole, have been developed to effectively treat this infection. However, these drugs are not effective in treating some cases because some isolates of this organism have developed a mechanism called mutations that enables them to become resistant to the drugs.
Understanding the cause of this resistance will help health professionals combat this challenge. Hence, many researchers have studied many causes of this resistance but have not sufficiently understood all possibilities. In this study, we used a variety of proven methods to discover a new cause of fluconazole resistance by Candida found in women and dogs. Our results showed that the resistant strain of the organism altered its DNA in a way that affected two vital anchors of the docking area for the drug. Consequently, the drug was rendered ineffective. This knowledge will arm pharmaceutical and medicinal chemists with information to create new designs that fit into such an abridged version of the target or redirect to other more stable spots that are potential drug targets within the organism.
Introduction
The incidence and prevalence of fungal infections have increased since the 1980s, especially in the population of immunocompromised patients.1–4 One of the most common Candida infections is vulvovaginal candidiasis, with symptoms that include vulvovaginal itching, friction and pain during sexual intercourse, inflammation, soreness in the vagina, burning discomfort when urinating, vaginal discharge, and vaginal cracks.5–7 According to a survey, vulvovaginal candidiasis affects 70–75% of women of childbearing age at least once in their lifetime,8 and 40–50% will experience reoccurrence.5 More than 17 different Candida species are known to be etiological agents of human fungal infection, and about 90% of invasive Candida infections are caused by C. albicans, C. glabrata, C. Parapsilosis, C. tropicalis, and C. krusei.4,9,10 The predominant cause of invasive fungal infection is C. albicans, of which the clinical indicators vary from superficial mucosal infection to deep invasive organ or systemic infection.11
Despite the progress made in the treatment of Candida infections, recent epidemiological studies indicate that the increasing incidence of antifungal resistant isolates of Candida in clinical settings poses a great challenge worldwide, which necessitates the search for new therapeutics.12–15 Within the limited range of antifungal agents available, the azoles, often referred to as the “workhorse” of antifungal treatment, have been the most frequently used option for Candida infections for decades.16–20 This is due to their effectiveness, relatively low cost, limited toxicity, and oral availability for systemic antifungal treatment. The azoles are fungistatic11,16 to Candida, hence lengthy, repeated, and inconsistencies with prophylaxis and treatment courses are the most probable causes of azole resistance in clinical isolates. Azoles work by inhibiting the biosynthesis of ergosterol,21 an indispensable component for maintaining the fluidity in the membranes of eukaryotic cells, which leads to the toxic accumulation of its precursor, lanosterol. One of the most commonly prescribed azole antifungals used for the treatment of C. albicans infections is fluconazole.22–24 Fluconazole inhibits the enzyme sterol 14–α demethylase (CYP51), which is essential for the biosynthesis of the fungal–specific membrane ergosterol.25,26 C. albicans CYP51 (CaCYP51) whose normal substrate is lanosterol, catalyzes the demethylation reaction of lanosterol in a three-step process towards producing ergosterol. Azole antifungal drugs inhibit this enzyme by binding their nucleophilic N–4 atom to the enzyme’s heme Fe (iron) at the sixth coordinate position, hence occupying the binding pocket competitively.27–29 CaCYP51 is encoded by the ERG11 gene,30 and altering the nature of the enzyme at the sequence level is a common strategy for evading the inhibitory action of the azoles, leading to resistance.27,31 Earlier reports have linked ERG11 mutations to three main amino acid sequence hot spots, namely 105–165, 266–287, and 405–488.32,33 In accord, Kumar et al reported an isolate with triple alterations (Y132C, F145L, A114V), of which Y132 and F145 sites were previously linked to azole resistance.34 They also discovered that one of the isolates had a novel mutation, G129R. In this study, we combined efficacy studies with advances in sequence analysis to characterize the mutational and polymorphic signatures in the ERG11 gene of clinical isolates of C. albicans strains with varying susceptibilities to fluconazole, isolated from women and dogs of reproductive age.
Materials and Methods
Sources of Candida Isolates
Candida isolates were obtained from freshly cultured high vaginal swabs (HVS) samples banked at the repository of the University of Nigeria Medical Center, Nsukka (UNNMC). These samples were collected from anonymous women of reproductive age, between the ages of 20 and 35 years, with symptoms of vulvovaginal candidiasis. Similarly, HVS samples from dogs (1–2 years) were obtained from the repository of the University of Nigeria Veterinary Teaching Hospital, Nsukka (UNVTH). A total of 57 human HVS (h–HVS) and 7 dog HVS (d-HVS) samples were collected, respectively.
Culture Condition and Isolation of Candida Strains
The HVS samples obtained were screened for yeast strains by streaking on Sabouraud Dextrose Agar (SDA),35,36 Thermo Scientific™ Oxoid™(40 g/L dextrose, 10 g/L peptone, 20 g/L agar, pH 5.6). The SDA was prepared according to the manufacturer’s instructions. To process the specimen, direct streaking with an inoculating loop was used to inoculate samples on SDA. Plates were incubated at 25–30°C in an inverted position for 24–48 h and monitored for growth.
Microscopic Identification and Characterization of Isolates
Isolates were phenotypically identified based on their microscopic and macroscopic characteristics as described by classical methodologies, including colony formation, and the production of germ tubes.37–39 The clinical samples were incubated in normal saline for 2 to 3 h at 30°C to 37°C. Samples were observed on slides under a light microscope (MoticTM, USA) and viewed for the short, slender, tube-like structures (germ tube). The presence of germ tubes is indicative of fungal species.
Differential Identification and Characterization of Candida spp. Using Chromogenic Media
The fungal–positive isolates were characterized using differential (Chromogenic Agar) media for Candida;40–43 Chromogenic Candida Agar (CHROMagar®; Paris, France).44–47 The agar plates were brought to room temperature before use. Inoculation was done by streaking the sample onto the plates. The plates were incubated in aerobic conditions at 30–37°C for 48 h and the different species of Candida identified by their colors.44–47
Antifungal Drug Susceptibility Testing
The susceptibility of the Candida isolates to the selected antifungal agent (fluconazole) was evaluated using disk diffusion test (DDT)48,49 and minimum inhibitory concentration (MIC),50 following guidelines of the Clinical and Laboratory Standards Institute (CLSI) document M27–A316.51,52 Suspension of inoculums were prepared in 5 mL of sterile saline (0.85%) and the turbidity was adjusted to 0.5 McFarland standards.53–58 Within 15 minutes of adjusting the turbidity, each isolate was inoculated onto a dried surface of prepared Mueller–Hinton agar plates using sterile cotton swabs. Antimicrobial disks containing 25 μg of fluconazole (Thermo ScientificTM; OxoidTM, USA) were dispensed and pressed firmly onto the inoculated agar plate’s surface to ensure complete contact with the agar surface. Plates were incubated at 37°C for 24 and 48 h and afterward examined. The inhibition zones were measured in millimeters and the results were interpreted using validated CLSI interpretive breakpoints for fluconazole in vitro susceptibility testing.55–58 Candida species have been classified as susceptible S (zone diameter ≥19 mm), susceptible dose-dependent SDD (15 to 18 mm), and resistant R (≤14 mm). The susceptibility data were acquired in triplicates.
DNA Isolation
ZYMO RESEARCH Fungal/Bacterial DNA Miniprep Kit was used to isolate the Candida DNA following the manufacturer’s instructions. Beta–mercaptoethanol was added to the Genomic Lysis Buffer to a final dilution of 500 μL per 100 mL. In a ZR Bashing BeadTM Lysis tube, 50–100 mg (wet weight) of Candida cells were weighed into 200 μL of water, followed by 750 μL of lysis solution, and centrifuged at 10,000 x g for 6 minutes. The supernatant was transferred to a Zymo–Spin™ IV Spin Filter in a collection tube and centrifuged further at 7000 x g for 1 minute. Genomic Lysis Buffer (1200 μL) was added to the filtrate in the collection tube. About 800 μL of the mixture was transferred to a Zymo–Spin™IIC Column in a collection tube and centrifuged at 10,000 x g for 1 minute. The Zymo–SpinTM IIC Column was loaded with 200 μL of DNA Pre–Wash Buffer and centrifuged at 10,000 x g for 1 minute. The Zymo–SpinTM IIC Column was loaded with 500 μL of g–DNA Wash Buffer and centrifuged for 1 minute at 10,000 x g. The Zymo–SpinTM IIC Column was transferred to a clean 1.5 mL microcentrifuge tube, 100 μL of DNA Elution Buffer was added directly to the column matrix, and the DNA was eluted by centrifugation at 10,000 x g for 30 seconds. The pure DNA was stored at –20°C for further use.
Molecular Identification of Candida Species by PCR–RFLP
Molecular identification of Candida species by PCR–RFLP based on the ribosomal region consisting of internal transcribed spacers ITS1 and ITS2 as well as the 5.8S gene is often used as a confirmatory test for the pure isolate of the organism (Supplementary Table 1).59 Amplification was performed in 50 μL volume of 25 μL of 2X PCR Master Mix, 1 μL forward and reverse primers (10 μM each), 1 μL of DNA template (40 ng/μL) and 28 μL water. The universal primer sequences were: ITS1– 5’–TCCGTAGGTGAACCTGCGG–3’, and ITS4– 5–TCCTCCGCTTATTGATATGC–3’. The samples were run on a 96–well Thermal Cycler (Thermo Fisher Scientific) as follows: 95°C (5 min), 95°C (30s), 56°C (30s), 72°C (30s), 37 cycles (step-2 to step-4), and a final extension at 72°C (5 min). Amplicons from the reaction were resolved on 1.5% agarose gel stained with ethidium bromide and analyzed using electrophoresis. A volume of 10 μL amplificons of the ribosomal region were digested with MspI restriction enzyme at 37°C for 2 h. The fragments from the digest were resolved on 2% agarose gel stained with ethidium bromide and analyzed using electrophoresis. The gel was read on BioDoc–It Imaging System (CA, USA). Purity of DNA was verified by amplifying the human beta–globin gene60 using the primers PCO3: 5’–ACACAACTGTGTTTCACTAGC–3’ and PCO5: 5’–GAAACCCAAGAGTCTTCTCT–3’.
PCR Amplification of the ERG11
The PCR reaction mixture (25 µL total volume) contained 12.5 µL PCR master mix, 1 μL forward and reverse primers (10 μM each), 1 μL of DNA template (40 ng/μL), and 10.5 μL deionized water. PCR was carried out using two pairs of primers that span the ERG11open reading frame: F1 (5′–AT GGCTATTGTTGAAACTGTCATT–3′), R1 (5-′GGATCAATATCACCACGTTCTC–3′); F2 (5′–ATTGGAGACGTGATGCTGCTCAA–3′), R2 (5’–TTAAAACATACAAGTTTCTCTT–3’). The PCR was performed in a 25–well thermocycler (Eppendorf, Germany). The amplification program for all reactions was as follows: initial denaturation at 94°C (5 min), denaturation at 94°C (30 s), annealing at 55°C (40 s), extension at 72°C (50 s), 30 cycles (step-2 to step-4) followed by final extension at 72°C (10 min). The PCR products were resolved on 2% agarose gel electrophoresis to assess their quality and integrity.
Sequencing the ERG11 Gene
PCR products were cleaned using ExoSAP according to the manufacturer’s protocol. Briefly, the Exo/SAP master mix was prepared by adding 50 µL Exonuclease I (NEB M0293) 20U/ul and 200 µL Shrimp Alkaline Phosphatase (NEB M0371) 1U/ul to a 0.6mL microcentrifuge tube. This was followed by mixing 10µL of PCR Mixture and 2.5µL Exo/SAP Mix. The mixture was properly mixed and incubated at 37°C for 30 min. The reaction was then stopped by heating the mixture at 95°C for 5 min. Gene sequencing was done with the ABI V3.1 Big dye kit following the instructions stipulated in the manufacturer’s manual. The labelled products were cleaned with the Zymo Seq clean–up kit following the manufacturer’s instruction. The cleaned products were injected on the ABI3500XL analyzers with a 50 cm array, using POP7.
Sequence–Level Investigation of the Resistance–Inducing Genetic Variants
In order to understand the observed variability in the pattern of resistance and susceptibility to fluconazole exhibited by the clinical isolates of interest, we employed several structural analytic approaches, which include multiple sequence and structural alignment, in silico site-directed mutagenesis and protein–ligand interaction profiling. The reference sequences and 3-dimensional structures that were used for the purpose of this study were obtained from the National Center for Biotechnology Information (NCBI)61 and Protein Data Bank (PDB),62 respectively. XM_711668.2 represents the accession number of the wild–type C. albicans lanosterol 14–α demethylase (CYP51) nucleotide sequence, while 5FSA and 4WMZ represent the PDB codes for both C. albicans and S. cerevisiae CYP51 proteins, in complex with Posaconazole and fluconazole, respectively.63,64 To study the variations in the nucleotide composition of the C. albicans drug-resistant and susceptible strains, different multiple sequence alignments (MSA) were conducted using the Clustal Omega software.65 The same platform was used for the comparative amino acid sequence of the C. albicans and S. cerevisiae CYP51 in an attempt to evaluate the degree of conservation of the selected residues of interest, while the 3D structural alignment was carried out using the alignment/superposition function of the pymol plugin manager.66 In addition, the C. albicans CYP51 mutant was computationally designed using the Chimera–curated Dynameomics rotamer library.65 All interactions were predicted using the protein–ligand interaction profiler,67 while saved outputs were visualized using the Pymol molecular visualizer.66
Molecular Dynamics Simulation
Using the Charmm36 force field and the 2019 version of GROMACS,68,69 we conducted molecular dynamics simulations on the wild-type and mutant C. albicans CYP51 in complex with their respective prosthetic heme. Solvation was carried out on both systems using the SPC (simple point charge) water molecules, and system neutralization were achieved with the inclusion of adequate chloride and sodium ions. A 5000 step steepest descent was used for the energy minimization of the systems and equilibration was achieved at 1 bar NPT and 300K NVT. With a production run of 100 ns, the comparative analysis of the conformational stability of both systems was estimated in reference to the output of equilibrium properties such as the RMSD (root mean square deviation), RMSF (root mean square fluctuation), Rg (radius of gyration), SASA (solvent accessible surface area), and the intramolecular hydrogen bonding.68,69
Sequence Retrieval and Phylogenetic Analysis
Additional nucleotide sequences of CandidaERG11gene were retrieved from NCBI–GeneBank61 for phylogenetic analysis. The criteria for selecting the additional 16 sequences are that they are closest to the four isolates we studied based on NCBI BLAST search, with a cut–off similarity score of 95% and 100% query coverage, they have 1587 bp coding sequence length, they only differ from the wild-type by at least a point mutation, and have been implicated in azoles susceptibility studies. The construction of a phylogenetic tree to depict the evolutionary relationship between the Candida strains based on the ERG11 sequence was carried out using Geneious Prime software70 and Molecular Evolutionary Genetic Analysis Software 7.0 (Mega7).71 The analysis was done using the Maximum Likelihood method (–10997.03) and the Tamura–Nei model with 1000 bootstrap replications.
Statistical Analysis
Other statistical analysis and building of graphs and charts were done using GraphPad Prism version 8.1.0 (221).
Results
Identification and Characterization of Candida Species
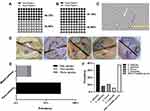
Approximately half of the h-HVS samples grew on SDA with a germ tube, indicating fungal presence (Figure 1A and C; Supplementary Table 2), and a similar result was obtained for d-HVS samples (Figure 1B and C; Supplementary Table 2). After considering the phenotypic characteristics of the dog samples, such as growth rate, color, and colony formation (observation not shown), the d-HVS–001 sample showed the right characteristics that necessitated its pick for further studies. We further characterized the Candida isolates at the species level, which was identified by varying colors on chromogenic (differential) media (Figure 1D), and the result showed different species of Candida, including C. albicans, C. tropicalis, C. krusei, C. glabrata, and other unidentified species.44–47 Some displayed mixed cultures, while others were pure cultures. The result of the study showed mostly pure cultures and a relatively lower percentage of mixed cultures (Figure 1E). Species-level screening of the 38 pure isolates obtained from the h-HVS showed a predominantly C. albicans species, followed by C. glabrata, C. krusei, and C. tropicalis (Figure 1F). The result further showed that the percentage of other unidentified species was as high as 37.84%. We identified the isolate from the dog (Can Iso–dog/029) as a C. albicans strain.
Isolates Exhibited Varying Susceptibility to Fluconazole
Antifungal susceptibility testing on the h-HVS samples revealed that the majority of the Candida isolates were susceptible (≥19 mm) to 25 g fluconazole, with a lower proportion being resistant (≤14 mm) and SDD (15–18 mm) strains (Table 1; Figure 2A–G). As shown in the result, the isolate from dog was susceptible. Can Iso–001, a C. albicans strain, was the most resistant (0 mm), and Can Iso–028, also a C. albicans strain, was the most susceptible. We extended the drug exposure time to 48 hours (Table 1), and compared it with the outcome at 24 hours. However, the result showed that there was no significant difference (p > 0.05) between the zone of inhibition and MIC values exhibited by the Candida isolates on 25 μg fluconazole at 24 h and 48 h (Figure 2H and I, respectively).
 |
Table 1 Susceptibility of Candida Isolates to 25 μg Fluconazole |
Disparities in the ERG11 Gene Sequence Elucidates the Varying Susceptibility to Fluconazole
The ERG11 gene codes for the Candida protein lanosterol 14–α demethylase (CYP51).72 Four (4) isolates from the experimental isolates, in addition to SC5314 were the strains of interest for ERG11 gene sequencing, including the most resistant (Can Iso–001), SSD (Can Iso–17), the most susceptible (Can Iso–028) and the dog isolate (Can Iso–029/dog). The nucleotide sequences of the ERG11 gene of the various samples showed that the primary sequence structure varied. It has been previously reported that the inhibition of fungal CYP51 by azoles occurs through the formation of an axial coordination bond between the azole drugs and the prosthetic heme iron of the protein, thereby affecting the reduction potential of the iron moiety.63 To further demonstrate this as related to this study, we analyzed the binding modes and interactions of two azole drugs (posaconazole and fluconazole) that have been complexed with the binding pocket of CYP51 in C. albicans and S. cerevisiae, respectively. A keen study of the output by the protein–ligand interaction profiler revealed various forms of interaction between the bound azole drugs and different residues of the CYP51 binding pocket and, most importantly, with the prosthetic heme iron (Figure 3A and B; Supplementary Tables 3 and 4).72
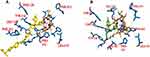 |
Figure 3 A depiction of the binding poses of two different azole drugs in the CYP51 drug-binding pocket. (A) shows the interaction of posaconazole (yellow) with different active site residues (blue) and prosthetic heme group (brown) of the C. albicans CYP51. (B) shows the interaction of fluconazole (green) with different active site residues (blue) and prosthetic heme group (brown) of the S. cerevisiae CYP51. Notes: A similar study by Zhang et al serves as additional evidence/support for our original analysis.72 |
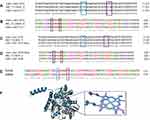
Several studies have reported that the prosthetic heme group of the fungal CYP51 protein is anchored by different active site residues that when mutated have the potential to disrupt the position of the heme, hence conferring drug resistance on the mutated fungal species.72–79 Based on this knowledge, we conducted a MSA between the nucleotide sequence (ERG11) of the wild-type C. albicans CYP51 protein and the nucleotide sequences of the mutated clinical isolates. Different levels of nucleotide and protein sequence similarity studies were conducted in this regard, in order to separate the resistant strains from the susceptible ones. The first alignment was directed towards the detection of specific mutations in the nucleotide sequences that code for the heme-binding amino acid residues in the C. albicans CYP51 active site. This was conducted on the assumption that mutations in the drug-binding site residues of CYP51 that reduce the stability of the prosthetic heme group affect the binding affinity of antifungal azole drugs, thus conferring resistance to various degrees. We aligned the nucleotide sequence of the wild-type CYP51 (XM_711668.2) with the sequences of the most resistant and SDD clinical isolates (Can–iso–001 and Can–iso–017, respectively). The results of this alignment study revealed two mutated codons (LEU–370 and PRO–375) that code for heme–binding residues in the active site of C. albicans CYP51 (Figure 4A and B; Supplementary Figures 1 and 2). We, therefore, speculated that the observed differences in the degree of resistance by both strains are linked to the mutation of these two residues to SER–370 (in both strains) and HIS–375 (only in Can–iso–001), as revealed in the amino acid sequence alignment.
We then asked if the nucleotide sequences from the susceptible strains exhibited any of the mutations that were observed in the resistant strains. For this reason, a separate MSA was conducted to include only nucleotide sequences of the wild-type and the susceptible strains of the C. albicans CYP51 (Can–iso–028 and Can–iso–dog). Our observation, which supports the earlier speculation, shows that the mutations of the CTC and CCA codons are directly linked to the drug-resistant attributes of the Can–iso–001 and Can–iso–017 isolates, as neither of the two susceptible strains (Can–iso–028 and Can–iso–dog) exhibited these mutations (Figure 4C and D; Supplementary Figures 3 and 4).
Additionally, we performed a pairwise alignment between the amino acid sequences of both C. albicans and S. cerevisiae CYP51 proteins and confirmed the conservation of these heme-binding residues of interest (Figure 4E; Supplementary Figure 5). To better understand the dynamics of the mutations linked to drug resistance, we carried out a structural alignment study on the 3D structures of C. albicans and S. cerevisiae CYP51 accessed from the Protein Database (PDB). The structural alignment showed a high degree of similarity in the binding orientation of the prosthetic heme group in both proteins (Figure 4F, Supplementary Figure 6A and 6B).
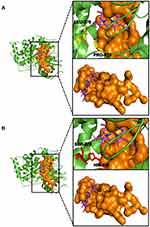
Using the Dynameomics rotamer library,80 we created an in-silico mutant of C. albicans CYP51 by replacing serine and histidine with LEU–370 and PRO–375, respectively. The result reveals a difference in the orientation of these heme-interacting residues in the wild-type (Figure 5A) and the mutant (Figure 5B). Additionally, this site-directed mutagenesis protocol was conducted in an attempt to also evaluate the effect of both mutations on the observed hydrophobic interaction with the prosthetic heme group. Upon the mutation of both residues to serine and histidine, respectively, the hydrophobic interaction between the wild-type residues (LEU–370 and PRO–375) and the prosthetic heme moiety was lost (Figure 6A and B). This interaction loss speculatively leads to the destabilization of the prosthetic heme, resulting in an eventual increase in resistance to the heme–binding antifungal azole drug (fluconazole).
The Resistance-Inducing Mutation Stabilized the CYP51 Protein at the Expense of the Prosthetic Heme
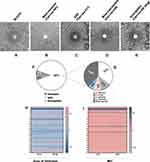
We used a comparative molecular dynamics simulation to study the movement of molecules and atoms within the wild-type and mutant C. albicans CYP51 proteins over a period of 100 ns after observing the interaction disparities caused by mutations affecting the prosthetic heme-binding pocket residues. This is to better understand their conformational stability changes. In molecular dynamics simulation, the molecular and atomic trajectories are determined through the numerical simplification of Newton’s equation of motion for systems of interacting particles.68,69 Forces between potential energies and their corresponding particles are usually estimated using molecular mechanics force fields or interatomic potentials.68,69 In this method, the RMSD measures the differences in the protein backbones from the initial to the final structural conformation.81,82 The protein conformation relative to its stability can be estimated through the observed deviations during the simulation. The larger the deviations, the less stable the protein structure.68,69 The RMSD plots showed that the wild-type protein exhibits a gradual increase in deviation up until 75 ns, after which the system stabilizes until the end of the simulation (Figure 7A). A different trajectory was observed in the mutant protein, which experienced a gradual increase in deviation till 30 ns, after which the system equilibrated and stabilized at an RMSD of 3 nm till the end of the simulation time. The regions of both proteins (wild-type and mutant C. albicans CYP51) that display a high degree of flexibility were also estimated using the residue-based RMSF. A comparative study of the RMSF plots showed that the mutant protein displayed fewer flexible residues compared to the wild-type (Figure 7B), indicating increased stability upon mutation.
The packing of amino acid residues is also known to have a great impact on the stability of a protein,83 and the measure of protein compactness has often been estimated using the Rg.84 We observed a lower value for the mutant protein’s radius of gyration (approximately 2.2 nm) compared to the wild-type (approximately 2.4 nm) (Figure 7C). This result is consistent with the outcome of the RMSD and RMSF, which indicates its increased stability over the wild-type. The low fluctuation as observed in the Rg plot was also maintained throughout the simulation. The compactness of a protein has also previously been defined as the ratio of its accessible surface area to the surface area of an ideal sphere of the same volume.85 Using the SASA plot, we estimated the solvent-exposed surface area of both the wild-type and mutant proteins.86,87 A comprehensive study of both plots reveals a lower degree of solvent accessibility upon mutation (Supplementary Figure 7), hence a higher degree of compactness and structural stability in comparison to the wild-type protein.
We also compared the RMSD trajectories of the bound prosthetic heme of the wild-type and mutant proteins. The wild-type heme equilibrated at about 60 ns with an approximate RMSD of 3 nm, unlike the mutant heme that maintained constant fluctuations all through the simulation period (Figure 8A). Protein-ligand hydrogen bonding provides specificity and directionality of interaction, which is a crucial molecular recognition aspect.88 While the wild-type heme ligand exhibited a drastic increase in hydrogen bonds throughout the simulation, the mutant heme experienced a drastic reduction in hydrogen bonds from 60 ns till the end of the simulation period (Figure 8B). This is in contrast to the wild-type CYP51 protein, which displays less hydrogen bond stability when compared to that of the mutant (Supplementary Figure 7). This result, supported by the ligand RMSD output, both suggest a reduction in the stability of the bound prosthetic heme upon the mutation of the active site residues (LEU–370 and PRO–375).
Evolutionary Relatedness of the Candida Strains Based on the ERG11 Gene Sequence
We analyzed the ancestrally unrooted phylogenetic relationship between 22 different C. albicans isolates based on the ERG11 gene sequence, and an additional ERG16 sequence that came up as a significant hit to all the ERG11 sequences of our isolates in this study (Figure 9). These strains include XM_711668 (SC5314), Can–iso–001, Can–iso–017, Can–iso–028, Can–iso–029, and 16 other pre-selected isolates from previous studies deposited in the GenBank, as well as the C. albicans ERG16 sequence (Supplementary Figure 8; Supplementary Table 5). The study showed that the new isolates studied here are categorically distinct from the other isolates analyzed. Our findings also showed that the most resistant isolate, Can–iso–001, is most closely related to the susceptible isolate from a dog (Can–iso–029). Our results further showed that the most related azole-resistant isolate to our isolates is HM194202.1 (SZ169) isolated in China, with the following nucleotide changes; T693A, C341T, and G1400R (a non–specific nucleotide encoded by the R). The least related isolate is AF153849.1 (J913004/1) isolated in Belgium, with the following sequence changes; A348T, C383A, C1355T, and A1390G. All these spots are different from the drug resistance-related position discovered in this work. We also conducted an XM_711668 (SC5314) ancestrally rooted phylogenetic analysis of the same set of sequences. We found that Can–iso–001, Can–iso–017, Can–iso–028, and Can–iso–029, are distant from SC5314 by 9 strains but are closer to the SC5314 than seven 7 strains of the 16 additional strains (Supplementary Figure 9).
Discussion
This study investigated the species distribution, azole susceptibility, ERG11 gene profile, implications for the CYP51 protein properties, and the molecular phylogenetic relationship (based on the ERG11 sequence) of Candida strains isolated from the vagina of women and dogs of reproductive age. From the h-HVS samples, we found out that about half of the patients showing symptoms similar to those of vaginal candidiasis do not have Candida infection as the swab from such showed no growth or germ tube typical of Candida (Figure 1A and C). A similar observation was obtained for the d-HVS samples (Figure 1B and C). This implies that other infections may display the same symptoms as those exhibited in vulvovaginal candidiasis. Some of the samples showed mixed cultures, while others showed pure cultures. Different species of Candida, including C. albicans, C. tropicalis, C. krusei, and C. glabrata, were identified in this study (Figure 1D), and many other isolates could not be identified by the differential method employed. We showed that most of the samples from women that had Candida infection had pure cultures and a lower number of mixed cultures, not exceeding three species (Figure 1E). A similar study identified mixed cultures in h-HVS samples,89 however, the study did not quantify how many isolates are in mixed cultures. Mixed cultures can further complicate drug efficacy, as slight molecular variations may exist between the different species at the drug target site.
Species characterization of our isolates by differential media showed C. albicans to be the predominant species, followed by C. glabrata, C. krusei, and C. tropicalis (Figure 1F). The proportion of the isolates that were unidentified NAC species was equated with the proportion of C. albicans. The relationship between the C. albicans and the other unidentified NAC species may be difficult to explain as the method of identification used in this study is limited. The only successful isolate from the dog was identified as a C. albicans strain. Other studies have reported a similar distribution as seen in our study. For example, Khadka et al reported 56% C. albicans (as the predominant species), 20% C. tropicalis, 14% C. glabrata, and 10% C. krusei in clinical isolates from various specimens including HVS, urine, blood, sputum, tracheal tubes, and catheter tips in a hospital in Nepal.90 Likewise, related studies also reported a comparable prevalence to our findings.91,92 In contrast to our study, which showed that C. glabrata is the most predominant non–albicans (NAC) species, a study reported that C. tropicalis was the most prevalent NAC species in clinical isolates obtained from a hospital in Hyderabad.93 Combining the prevalence of the other unidentified NAC species and the NAC (identified) species, we show that NAC has a greater prevalence than C. albicans, and other studies have corroborated this observation,94,95 making the emergence of NAC species of important concern. The most predominant species in mixed culture with C. albicans were other unidentified NAC species, explaining the substantial thriving of C. albicans along with other species (Table 1). This indicates that there are diverse Candida species that are harbored in the vagina of women apart from the common species such as C. albicans, C. krusei, C. glabrata, C. tropicalis, and C. parapsilosis. The significance of this finding is that other unidentified Candida species, such as the recently trending C. dubliniensis,92 for which resistance to fluconazole has also been identified96–98 may substitute for C. albicans under favorable conditions.
Antifungal susceptibility testing of Candida is largely recognized as a useful aid in optimizing the treatment of Candida infections.51,52,57 This is essential due to the emergence of resistant strains that continuously threaten azole therapy as recently reported in a surveillance study on fluconazole resistance in both albicans and NAC species.99 Fluconazole is often the first choice for treating vaginitis in many parts of the world.100–104 In our study, the antifungal susceptibility testing on human isolates revealed that most of the Candida isolates were susceptible to 25 μg fluconazole (Table 1 and Figure 2). This observation shows that fluconazole remains a very useful antifungal drug for Candida infection. Overall, only about 5% of the isolates from humans investigated were resistant to fluconazole, and about 5% were SSD. This low percentage suggests that the need for new antifungals for Candida infection, though very important, may not be pressing currently. The SDD strains could also become susceptible by increasing the dosage or usage frequency of fluconazole.105 Our results showed that the most resistant isolate, a C. albicans strain, was completely resistant to fluconazole. We speculated that this strain may have undergone mutations as well as evolutionary adaptation in the ERG11 gene such that it has countered all possible mechanisms of fluconazole action.24,26,27,31,75 However, other resistance mechanisms (not investigated in this present study) apart from mutations in ERG11 may also be responsible.106–108 We extended the duration of the susceptibility testing from 24 h to 48 h to check for any significant improvement in efficacy.109,110 Our findings showed that there was no significant difference (p > 0.05) between the zone of inhibition at 24 h and 48 h (Figure 2H and I, respectively).
Four (4) isolates, in addition to the SC5314 strain, were the strains of interest selected for ERG11 gene sequencing and profiling, comprising of the most resistant isolate (Can Iso–001), an SDD isolate (Can Iso–17), the most susceptible isolate (Can Iso–028), and the dog isolate (Can Iso–029). The sequence analysis of the ERG11 gene for the various isolates showed that the nucleotide compositions of each strain varied. Differences observed between sequences of strains from the same species could be attributed to mutations or single nucleotide polymorphism (SNPs), and may also be due to changes resulting from counter mechanisms against antifungal agents, as the case may be.111–113 Variations in the ERG11 gene sequence have very diverse effects on the corresponding protein properties, including stability, electrostatic effects, protein packing, local and global structure, interactions, abundance, catalytic function, protein recognition, transport function of protein and other properties.111–113 The effects on protein tertiary structure further includes effect on protein dynamics, which covers those affecting allosteric sites, structural disorder, induced fit, and structural flexibility of the enzyme.114–116 Many deleterious variants, such as large deletions, protein truncations, and amphigoric amino acid insertions, are straightforward to explain. Usually, the most difficult ones are minor sequence alterations, most often resulting in amino acid substitutions in the protein. Therefore, variations in primary, secondary, and tertiary structures of CYP51 proteins (encoded by ERG11) are more difficult to explain than those of the ERG11 DNA sequence itself.
Previous studies have reported several mutations in the ERG11 gene of C. albicans that are linked to resistance to fluconazole and other azoles–based antifungal drugs in clinical isolates.27,33,117,118 The exploration of these mutations and their impacts could therefore provide major insights into the mechanisms that underlie their resistance to the azole drugs. In–silico techniques in the modern era play a crucial role in our understanding of biological systems.119 As part of these techniques, mutations are often generated in the proteins in order to elucidate the effects on the stability and conformation of the tertiary or quaternary structures.82 These techniques are currently used in the prediction of different biological mechanisms, such as protein–protein interactions (PPI), DNA-protein interactions, and drug resistance and sensitivity.82 The substitution of different amino acids in the C. albicans CYP51 protein is one of the most critical mechanisms that contribute to azole drug resistance in the organism.27,33,117,118 Studying the effect of residue substitution in this protein is therefore crucial to the identification of important residues that may be involved in the resistance of C. albicans to drugs.
Fluconazole has been used for more than 30 years as the major therapeutic agent for the treatment of C. albicans–linked infections, and there is substantial data available on clinical isolates of C. albicans that have displayed varying degrees of resistance to the drug.12,13,74,75,106,108,118 Various mechanisms of acquired resistance towards antifungal azoles have been proposed, and mutations in CYP51 are listed among such mechanisms, although their contribution to the resistance phenomenon remains inadequately understood.120 Considering the close structural evolutionary similarity of the CYP51 family across fungal species, and the property of the protein towards the preservation of their conserved biological function,121–124 it is unlikely to experience a high mutational frequency in the protein. Earlier studies have summarized that most of the reported mutations in the CYP51 from strains of C. albicans that displayed resistance against fluconazole have also been reported in susceptible strains, possibly making them irrelevant to the mechanism of resistance.120 Morio et al reported 10 mutations in the C. albicans CYP51 structure, which has so far not been detected in sequences of susceptible strains and 5 of these mutations, which include Y132H, Y132F, K143R, G307S, and S405F were reported as part of the exposed residues in the drug-binding pocket of the protein, suggesting that they might potentially affect the binding affinity of fluconazole. In agreement with our protein–ligand interaction prediction report (Figure 3A and B; Supplementary Tables 3 and 4), Morio et al reported that 2 of the 5 listed active site residues of the C. albicans CYP51 structure (TYR–132 and LYS–143) form hydrogen bond interactions with the bound prosthetic heme group. Abolishing the hydrogen bond interaction or changes in the hydrogen bond strength as a result of the mutation of both residues to histidine and arginine, respectively, are suggested to have a possible effect on the heme iron redox potential;120 thereby affecting the ability of the iron to coordinate the basic nitrogen of the azole ring.120 Several mutations (spontaneous or SNPs) were observed in the nucleotide sequence of the four clinical isolates that were used in this study, but as a result of the reported relevance of the heme-binding active site residues to the drug resistance attribute of the C. albicans CYP51,29,74,77–79,116 our study was targeted at these set of residues in order to investigate the impact of mutations on the observed variability in resistance and susceptibility to fluconazole.
Through MSA protocols, we detected two codons in the nucleotide sequence of the wild-type (CTC and CCA) that codes for LEU–370 and PRO–375. An alignment of the wild-type sequence (XM_711668.2) with sequences of the resistant clinical isolates (Can–iso–001 and Can–iso–017) revealed that these residues were mutated to serine and histidine, respectively (Figure 4A and B). In order to validate the relevance of the observed mutations to drug resistance as previously reported, we repeated the alignment using the wild-type nucleotide sequence and the nucleotide sequences of the drug-sensitive isolates (Can–iso–028 and Can–iso–dog). The output of this alignment (Figure 4C and D) showed that the codons for the heme-binding residues were not mutated in the susceptible strains, which further supports our observation. So far, the L370S and P375H resistance–linked mutations have not been previously reported in any literature. The alignment of their amino acid sequences and 3D structural alignment between the C. albicans and S. cerevisiaeCYP51 showed a high degree of conservation both generally and for the two residues of interest (LEU–370 and PRO–375) as well as a conserved binding mode of the prosthetic heme group (Figure 4E and F). To further elucidate the relevance of these residues to the observed resistance to fluconazole by the clinical isolates, the output from our site-directed mutagenesis study also revealed a loss of hydrophobic interaction between the residues of interest and the prosthetic heme group (Figure 5). In the wild-type caCYP51, LEU–370, and PRO–375 form part of the heme interacting residues however, the mutation in these residues to SER–370 and HIS–375, respectively, led to the loss of these residues in the interaction profile (Figure 6). This possibly leads to the heme moiety not being effectively held in place.120
More insight into the mechanism of resistance of the mutant was further revealed through the molecular dynamics simulation, involving the comprehensive analysis of the RMSD, RMSF, Rg, SASA, and the hydrogen bond plots of the wild-type and mutant protein–ligand interaction. The directional interaction that supports molecular recognition, protein folding, and protein structure is provided by hydrogen bonds.88 Most protein structural cores are made up of secondary structures, such as the alpha-helix and beta-sheet. This satisfies the potential of hydrogen bonding between the carbonyl oxygen of the main chain and the buried nitrogen in the protein hydrophobic core.88 The results of the molecular dynamics simulation collectively suggested that the novel mutant strain of the pathogen in order to resist the antifungal activity of fluconazole, mutated the LEU–370 and PRO–375 to ultimately alter the hydrogen bonding pattern, and stabilize CYP51 at the expense of the bound prosthetic heme (Figures 7 and 8; Supplementary Figure 7).
Environment could play a major role in the biological evolution of organisms in order to adapt and survive. This has led to highly divergent variations in genetic sequences within the same gene of the same species of organisms. Some of these genetic variations are associated with drug resistance. ERG11 gene, being a gene encoding a major protein (caCYP51) essential for the survival of Candida, we were interested in analyzing its evolutionary divergence in isolates that have been implicated in resistance. It will be important to understand the relatedness of the isolates from our environment to other isolates obtained in other regions. We included the criteria for the selection in the materials and methods section. The result of our unrooted phylogenetic analysis between our isolates of interest and other pre-selected isolates based on the ERG11 gene sequence revealed that the most resistant isolate Can–iso–001, is most closely related to the susceptible isolate from dogs (Can–iso–029). This observation, though yet to be empirically proven, suggests that Candida strains could transfer between humans and animals via surfaces or other means of contact and assume different behaviors in different environments. We further showed that the most related azole-resistant isolate to our isolates is HM194202.1 (SZ169) isolated in China, and the least related isolate is AF153849.1 (J913004/1) isolated in Belgium with different mutation spots in their ERG11 sequence. This strongly supports the knowledge that other parts of the caCYP51 protein can determine the susceptibility to azoles, and that our variants of the resistant isolates are new. These findings add to the knowledge that multiple regions of the CYP51 protein either singly, or in combination can determine the susceptibility of Candida strains to azoles.
Conclusion
This study attempted to characterize the species distribution, azole susceptibility, ERG11 gene, and caCYP51 protein profiles of clinical isolates of Candida from the vagina of women and dogs of reproductive age. We focused more on exploring the patterns in sequence variations in the ERG11 gene, and the consequences on the caCYP51 protein, that may determine the susceptibility and resistance of Candida strain to azoles. We achieved this by applying proven classical susceptibility testing methods and molecular sequence analysis techniques to the clinical isolates studied. The result further established that the novel mutant strain mutated the LEU–370 and PRO–375 to stabilize caCYP51 at the expense of the bound prosthetic heme in order to resist the antibiotic activity of fluconazole. Taken together, our result showed new mutations in the heme-binding pocket of caCYP51 that explains the resistance to fluconazole, exhibited by the Candida isolates.
Limitations of the Study
This study provided robust data to support our findings however, a few limitations are noted. First, the differential method for identifying the specific species of Candida is narrow, although highly efficient. Hence, the unidentified NAC species could not be sufficiently characterized. Secondly, other metabolic mechanisms, such as drug efflux not investigated here may also be contributory to the observed resistance to fluconazole however, this study focused on the mechanism of action of fluconazole at the drug target site. Additionally, reverse genetics targeting these points of interest is yet to be applied in order to rule out all other metabolic and genetic possibilities that could be contributory to the resistance.
Abbreviations
BLAST, basic local alignment search tool; caCYP51, Candida CYP51; DDT, disk diffusion test; d-HVS, dog high vaginal swab; h-HVS, human high vaginal swab; HVS, high vaginal swab; MIC, minimum inhibitory concentration; MSA, multiple sequence alignments; NAC, non-albicans; NCBI, National Center for Biotechnology Information; PDB, protein database; RMSD, root mean square deviation; RMSF, root mean square fluctuation; Rg, radius of gyration; SASA, solvent accessible surface area; SPC, simple point charge.
Data Sharing Statement
Nucleotide sequences used in this study are available at the National Center for Biotechnology Information (NCBI) database with the following Gene accession numbers, OL944653; OL944654; OL944655; OL944656; XM_711668.2; AF153849.1; AF153846.1; GQ202072.1; HM194171.1; HM194175.1; GQ202079.1; MG162590.1; EU819550.1; EU980038.1; KX631421.1; EU848302.1; KM609923.1; HM194203.1; FJ002303.1; X13296.1; KM609920.1; HM194202.1.
Ethical Approval and Consent
We observed the ethical guidelines for the use of humans and experimental animals in biomedical research in accordance with the Declaration of Helsinki, and approval was granted by the review board of the Ethics and Biosafety Committee, Faculty of Biological Sciences, University of Nigeria, Nsukka (UNN/FBS/EC/1079). Based on the research and clinical guidelines, informed consent was obtained from the study participants at the University of Nigeria Medical Center, Nsukka (UNNMC). Collection of the h-HVS and d-HVS samples was approved by the institutional heads of the University of Nigeria Medical Center, Nsukka (UNNMC), and the University of Nigeria Veterinary Teaching Hospital (UNVTH), respectively, from which the clinical isolates of Candida were obtained, respectively.
Acknowledgments
The authors thank the management of the University of Nigeria Medical Center, Nsukka, and the University of Nigeria Veterinary Teaching Hospital, Nsukka for their approval of samples for the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors funded the work as no external funding was provided.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jerez Puebla LE. Fungal infections in immunosuppressed patients. Immunodeficiency. 2012. doi:10.5772/51512
2. Rodrigues ML, Nosanchuk JD, Reynolds TB. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis. 2020;14(2):1–9. doi:10.1371/journal.pntd.0007964
3. Low CY, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep. 2011;3(1):1–8. doi:10.3410/M3-14
4. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi. 2017;3(4):57. doi:10.3390/jof3040057
5. Hurley R, De Louvois J. Candida vaginitis. Postgrad Med J. 1979;55:645–647. doi:10.1136/pgmj.55.647.645
6. Nyirjesy P. Chronic vulvovaginal candidiasis. Am Fam Physician. 2001;63(4):697–702. doi:10.1056/nejmp048152
7. Paladine HL, Desai UA. Vaginitis: diagnosis and treatment. Am Acad Fam Physicians. 2018;97(5):321–329.
8. Sobel JD. Vulvovaginal candidiasis. Lancet. 2007;369(9577):1961–1971. doi:10.1016/S0140-6736(07)60917-9
9. Musa K, Ahmed M, Shahpudin S, et al. Resistance of Candida glabrata to drugs and the host immune system. Clin Microbiol Infect Dis. 2018;3(3):1–4. doi:10.15761/cmid.1000145
10. Bhattacharjee P. Epidemiology and antifungal susceptibility of Candida species in a tertiary care hospital, Kolkata, India. Curr Med Mycol. 2016;2(2):20–27. doi:10.18869/acadpub.cmm.2.2.5
11. Gullo A. Invasive fungal infections. The challenge continues. Drugs. 2009;69(1):65–73. doi:10.2165/11315530-000000000-00000
12. Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi:10.1093/cid/ciw691
13. Alexander BD, Johnson MD, Pfeiffer CD, et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis. 2013;56(12):1724–1732. doi:10.1093/cid/cit136
14. Rodriguez L, Bustamante B, Huaroto L, et al. A multi-centric study of Candida bloodstream infection in Lima-Callao, Peru: species distribution, antifungal resistance and clinical outcomes. PLoS One. 2017;12(4):1–12. doi:10.1371/journal.pone.0175172
15. Maheronnaghsh M, Fatahinia M, Dehghan P, Teimoori A. Identification of Candida species and antifungal susceptibility in cancer patients with oral lesions in Ahvaz, Southern West of Iran. Adv Biomed Res. 2020;9(1):50. doi:10.4103/abr.abr_214_19
16. Perfect JR. Is there an emerging need for new antifungals? Expert Opin Emerg Drugs. 2016;21(2):129–131. doi:10.1517/14728214.2016.1155554
17. Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12(1):40–79. doi:10.1128/cmr.12.1.40
18. The fungus among us: an antifungal review. https://uspharmacist.com/article/the-fungus-among-us-an-antifungal-review.
19. Scheinfeld N. Ketoconazole: a review of a workhorse antifungal molecule with a focus on new foam and gel formulations. Drugs Today. 2008;44(5):369–380. doi:10.1358/DOT.2008.44.5.1216598
20. McKeny PT, Nessel TA, Zito PM. Antifungal Antibiotics. StatPearls; 2021.
21. Ahmad A, Khan A, Manzoor N, Khan LA. Evolution of ergosterol biosynthesis inhibitors as fungicidal against Candida. Microb Pathog. 2010;48(1):35–41. doi:10.1016/j.micpath.2009.10.001
22. Autmizguine J, Smith PB, Prather K, et al. Effect of fluconazole prophylaxis on Candida fluconazole susceptibility in premature infants. J Antimicrob Chemother. 2018;73(12):3482–3487. doi:10.1093/jac/dky353
23. Gong Y, Liu W, Huang X, Hao L, Li Y, Sun S. Antifungal activity and potential mechanism of n-butylphthalide alone and in combination with fluconazole against Candida albicans. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.01461
24. Rybak JM, Muñoz JF, Barker KS, et al. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio. 2020;11(3):1–16. doi:10.1128/mBio.00365-20
25. Warrilow AGS, Melo N, Martel CM, et al. Expression, purification, and characterization of Aspergillus fumigatus sterol 14-α demethylase (CYP51) isoenzymes A and B. Antimicrob Agents Chemother. 2010;54(10):4225–4234. doi:10.1128/AAC.00316-10
26. Warrilow AGS, Mullins JGL, Hull CM, et al. S279 point mutations in Candida albicans sterol 14-α demethylase (CYP51) reduce in vitro inhibition by fluconazole. Antimicrob Agents Chemother. 2012;56(4):2099–2107. doi:10.1128/AAC.05389-11
27. Flowers SA, Colón B, Whaley SG, Schuler MA, David Rogers P. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother. 2015;59(1):450–460. doi:10.1128/AAC.03470-14
28. De Petris A, Crestoni ME, Pirolli A, et al. Binding of azole drugs to heme: a combined MS/MS and computational approach. Polyhedron. 2015;90:245–251. doi:10.1016/j.poly.2015.02.011
29. Balding PR, Porro CS, McLean KJ, et al. How do azoles inhibit cytochrome P450 enzymes? A density functional study. J Phys Chem A. 2008;112(50):12911–12918. doi:10.1021/jp802087w
30. Warrilow AG, Parker JE, Kelly DE, Kelly SL. Azole affinity of sterol 14-demethylase (CYP51) enzymes from Candida albicans and homo sapiens. Antimicrob Agents Chemother. 2013;57(3):1352–1360. doi:10.1128/AAC.02067-12
31. Xiang MJ, Liu JY, Ni PH, et al. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013;13(4):386–393. doi:10.1111/1567-1364.12042
32. Marichal P, Vanden Bossche H, Odds FC, et al. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41(10):2229–2237. doi:10.1128/aac.41.10.2229
33. Marichal P, Koymans L, Willemsens S, et al. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145(10):2701–2713. doi:10.1099/00221287-145-10-2701
34. Kumar A, Nair R, Kumar M, et al. Assessment of antifungal resistance and associated molecular mechanism in Candida albicans isolates from different cohorts of patients in North Indian state of Haryana. Folia Microbiol (Praha). 2020;65(4):747–754. doi:10.1007/s12223-020-00785-6
35. Njunda AL, Nsagha DS, Assob JCN, Kamga HL, Teyim P. In vitro antifungal susceptibility patterns of Candida albicans from HIV and aids patients attending the Nylon Health District hospital in Douala, Cameroon. J Public Health Africa. 2012;3(1):4–7. doi:10.4081/jphia.2012.e2
36. Bhargava A, Dakwala F, Saigal S, Mehra S. Identification of Candida albicans by using different culture medias and its association in potentially malignant and malignant lesions. Contemp Clin Dent. 2011;2(3):188. doi:10.4103/0976-237x.86454
37. Matare T, Nziramasanga P, Gwanzura L, Robertson V. Experimental germ tube induction in Candida albicans: an evaluation of the effect of sodium bicarbonate on morphogenesis and comparison with pooled human serum. Biomed Res Int. 2017;2017:8–13. doi:10.1155/2017/1976273
38. Munin E, Giroldo LM, Alves LP, Costa MS. Study of germ tube formation by Candida albicans after photodynamic antimicrobial chemotherapy (PACT). J Photochem Photobiol B. 2007;88(1):16–20. doi:10.1016/j.jphotobiol.2007.04.011
39. Torosantucci A, Romagnoli G, Chiani P, et al. Candida albicans yeast and germ tube forms interfere differently with human monocyte differentiation into dendritic cells: a novel dimorphism-dependent mechanism to escape the host’s immune response. Infect Immun. 2004;72(2):833–843. doi:10.1128/IAI.72.2.833-843.2004
40. Houang ETS, Chu KC, Koehler AP, Cheng AFB. Use of CHROMagar Candida for genital specimens in the diagnostic laboratory. J Clin Pathol. 1997;50(7):563–565. doi:10.1136/jcp.50.7.563
41. Willinger B, Hillowoth C, Selitsch B, Manafi M. Performance of Candida ID, a new chromogenic medium for presumptive identification of Candida species, in comparison to CHROMagar Candida. J Clin Microbiol. 2001;39(10):3793–3795. doi:10.1128/JCM.39.10.3793-3795.2001
42. Merlino J, Tambosis E, Veal D. Chromogenic tube test for presumptive identification or confirmation of isolates as Candida albicans. J Clin Microbiol. 1998;36(4):1157–1159. doi:10.1128/jcm.36.4.1157-1159.1998
43. Odds FC, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32(8):1923–1929. doi:10.1128/jcm.32.8.1923-1929.1994
44. Lahoylahoy LD, Mendoza BC. Evaluation of CHROMagar Candida in the rapid identification of medically important species of Candida. Int J Biosci. 2017;11(1):380–385. doi:10.12692/ijb/11.1.380-385
45. Sharma P, Sambyal S, Shrivastava D. Evaluation of Candida species from clinical specimens by using chromagar. Int J Adv Res. 2017;5(2):1750–1755. doi:10.21474/ijar01/3337
46. Mathavi S, Sasikala G, Kavitha A, Priyadarsini RI. CHROMagar as a primary isolation medium for rapid identification of Candida and its role in mixed Candida infection in sputum samples. Indian J Microbiol Res. 2016;3(2):141. doi:10.5958/2394-5478.2016.00033.9
47. Faraz A, Ghaffar UB, Tahir Ansari WS. Evaluation of diagnostic efficacy of chromagar Candida for differentiation: Taylor’s libraries. ISRA Med J. 2016;8(4):5.
48. Barry AL, Brown SD. Fluconazole disk diffusion procedure for determining susceptibility of Candida species. J Clin Microbiol. 1996;34(9):2154–2157. doi:10.1128/jcm.34.9.2154-2157.1996
49. Yücesoy M, Şentürker Güldaş N, Yuluǧ N. Disk diffusion method for fluconazole susceptibility testing of Candida albicans strains. J Chemother. 2001;13(2):161–166. doi:10.1179/joc.2001.13.2.161
50. Kiraz N, Dag I, Oz Y, Yamac M, Kiremitci A, Kasifoglu N. Correlation between broth microdilution and disk diffusion methods for antifungal susceptibility testing of caspofungin, voriconazole, amphotericin B, itraconazole and fluconazole against Candida glabrata. J Microbiol Methods. 2010;82(2):136–140. doi:10.1016/j.mimet.2010.05.002
51. Clinical and Laboratory Standards Institute Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Vol. 28.
52. Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts.
53. Alyousef AA. Antifungal activity and mechanism of action of different parts of Myrtus communis growing in Saudi Arabia against. J Nanomater. 2021;2021. doi:10.1155/2021/3484125
54. Pam VK, Akpan JU, Oduyebo OO, et al. Fluconazole susceptibility and ERG11 gene expression in vaginal Candida species isolated from Lagos Nigeria. Int J Mol Epidemiol Genet. 2012;3(1):84–90.
55. Alastruey-Izquierdo A, Cuenca-Estrella M. EUCAST and CLSI: how to assess in vitro susceptibility and clinical resistance. Curr Fungal Infect Rep. 2012;6(3):229–234. doi:10.1007/s12281-012-0100-3
56. Cuesta I, Bielza C, Cuenca-Estrella M, Larrañaga P, Rodríguez-Tudela JL. Evaluation by data mining techniques of fluconazole breakpoints established by the Clinical and Laboratory Standards Institute (CLSI) and comparison with those of the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrob Agents Chemother. 2010;54(4):1541–1546. doi:10.1128/AAC.01688-09
57. Pfaller MA, Diekema DJ, Sheehan DJ. Interpretive breakpoints for fluconazole and Candida revisited: a blueprint for the future of antifungal susceptibility testing. Clin Microbiol Rev. 2006;19(2):435–447. doi:10.1128/CMR.19.2.435-447.2006
58. Rex JH, Pfaller MA, Galgiani JN, et al. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1997;24(2):235–249. doi:10.1093/clinids/24.2.235
59. Ortiz B, Pérez-Alemán E, Galo C, Fontecha G. Molecular identification of Candida species from urinary infections in Honduras. Rev Iberoam Micol. 2018;35(2):73–77. doi:10.1016/j.riam.2017.07.003
60. Erazo BM, Ramírez GA, Cerrato LE, et al. Prevalence of Hb S (HHB: c.20A>T) in a Honduran population of African descent. Hemoglobin. 2015;39(2):134–137. doi:10.3109/03630269.2015.1012294
61. Sayers EW, Beck J, Brister JR, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020;48(D1):D9–D16. doi:10.1093/nar/gkz899
62. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi:10.1093/nar/28.1.235
63. Hargrove TY, Friggeri L, Wawrzak Z, et al. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J Biol Chem. 2017;292(16):6728–6743. doi:10.1074/jbc.M117.778308
64. Sagatova AA, Keniya MV, Wilson RK, Monk BC, Tyndall JDA. Structural insights into binding of the antifungal drug fluconazole to Saccharomyces cerevisiae lanosterol 14α-demethylase. Antimicrob Agents Chemother. 2015;59(8):4982–4989. doi:10.1128/AAC.00925-15
65. Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. doi:10.1002/pro.3290
66. Faure G, Joseph AP, Craveur P, et al. IPBAvizu: a PyMOL plugin for an efficient 3D protein structure superimposition approach. Source Code Biol Med. 2019;14(1):1–5. doi:10.1186/s13029-019-0075-3
67. Adasme MF, Linnemann KL, Bolz SN, et al. PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021;49(W1):W530–W534. doi:10.1093/nar/gkab294
68. Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1–3):43–56. doi:10.1016/0010-4655(95)00042-E
69. Abraham MJ, Murtola T, Schulz R, et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi:10.1016/j.softx.2015.06.001
70. Geneious Prime. Geneious all rights reserved. molecular biology and sequence analysis software. Available from: https://www.geneious.com/prime/.
71. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi:10.1093/molbev/msw054
72. Zhang J, Li L, Lv Q, Yan L, Wang Y, Jiang Y. The fungal CYP51s: their functions, structures, related drug resistance, and inhibitors. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.00691
73. Liu M, Zheng N, Li D, et al. Cyp51A-based mechanism of azole resistance in Aspergillus fumigatus: illustration by a new 3D Structural Model of Aspergillus fumigatus CYP51A protein. Med Mycol. 2016;54(4):400–408. doi:10.1093/mmy/myv102
74. Sagatova AA, Keniya MV, Wilson RK, Sabherwal M, Tyndall JDA, Monk BC. Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14α-demethylase. Sci Rep. 2016;6:1–11. doi:10.1038/srep26213
75. Parker JE, Warrilow AGS, Price CL, Mullins JGL, Kelly DE, Kelly SL. Resistance to antifungals that target CYP51. J Chem Biol. 2014;7(4):143–161. doi:10.1007/s12154-014-0121-1
76. Becher R, Wirsel SGR. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl Microbiol Biotechnol. 2012;95(4):825–840. doi:10.1007/s00253-012-4195-9
77. Lamb DC, Kelly DE, Schunck WH, et al. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272(9):5682–5688. doi:10.1074/jbc.272.9.5682
78. Sun B, Huang W, Liu M. Evaluation of the combination mode of azoles antifungal inhibitors with CACYP51 and the influence of Site-directed mutation. J Mol Graph Model. 2017;73:157–165. doi:10.1016/j.jmgm.2017.02.009
79. Rosam K, Monk BC, Lackner M. Sterol 14α-demethylase ligand-binding pocket-mediated acquired and intrinsic azole resistance in fungal pathogens. J Fungi. 2021;7(1):1–22. doi:10.3390/jof7010001
80. Scouras AD, Daggett V. The dynameomics rotamer library: amino acid side chain conformations and dynamics from comprehensive molecular dynamics simulations in water. Protein Sci. 2011;20(2):341–352. doi:10.1002/pro.565
81. Kufareva I, Abagyan R. Methods of protein structure comparison in homology modeling: methods and protocols. In: Methods of Protein Structure Comparison in Homology Modeling: Methods and Protocols, Methods in Molecular Biology. Vol. 857. Humana Press; 2012:231–257. doi:10.1007/978-1-61779-588-6
82. Aier I, Varadwaj PK, Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep. 2016;6:1–10. doi:10.1038/srep34984
83. Lobanov MI, Bogatyreva NS, Galzitskaia OV. Radius of gyration is indicator of compactness of protein structure. Mol Biol (Mosk). 2008;42(4):701–706.
84. Lobanov MY, Bogatyreva NS, Galzitskaya OV. Radius of gyration as an indicator of protein structure compactness. Mol Biol. 2008;42(4):623–628. doi:10.1134/S0026893308040195
85. Tsai CJ, Nussinov R. Hydrophobic folding units at protein-protein interfaces: implications to protein folding and to protein-protein association. Protein Sci. 1997;6(7):1426–1437. doi:10.1002/pro.5560060707
86. Gromiha MM. Protein structure prediction. Protein Bioinform. 2010;1986:143–207. doi:10.1016/b978-8-1312-2297-3.50005-9
87. Fullerton GD, Cameron IL. Water compartments in cells. Methods Enzymol. 2007;428:1–28. doi:10.1016/S0076-6879(07)28001-2
88. Hubbard RE, Kamran Haider M. Hydrogen bonds in proteins: role and strength. Encycloped Life Sci. 2010. doi:10.1002/9780470015902.a0003011.pub2
89. Montes K, Ortiz B, Galindo C, Figueroa I, Braham S, Fontecha G. Identification of Candida species from clinical samples in a Honduran tertiary hospital. Pathogens. 2019;8(4):1–11. doi:10.3390/pathogens8040237
90. Khadka S, Sherchand JB, Pokhrel BM, et al. Isolation, speciation and antifungal susceptibility testing of Candida isolates from various clinical specimens at a tertiary care hospital, Nepal. BMC Res Notes. 2017;10(1):1–5. doi:10.1186/s13104-017-2547-3
91. Manikandan C, Amsath A. Characterization and susceptibility pattern of Candida species isolated from urine samples in Pattukkottai, Tamilnadu, India. Int J Pure Appl Zool. 2015;3(1):17–23.
92. Reddy Edula A. Antifungal susceptibility of clinically significant Candida species by disk diffusion method. IP Int J Med Microbiol Trop Dis. 2021;7(2):77–80. doi:10.18231/j.ijmmtd.2021.017
93. Swoboda-Kopec E, Kawecki D, Wroblewska M, Krawczyk M, Luczak M. Epidemiology and susceptibility to antifungal agents of fungi isolated from clinical specimens from patients hospitalized in the Department of General and Liver Surgery of the Medical University of Warsaw. Transplant Proc. 2003;35(6):2298–2303. doi:10.1016/S0041-1345(03)00757-7
94. Mokaddas EM, Al-Sweih NA, Khan ZU. Species distribution and antifungal susceptibility of Candida bloodstream isolates in Kuwait: a 10-year study. J Med Microbiol. 2007;56(PART2):255–259. doi:10.1099/jmm.0.46817-0
95. Pfaller MA, Andes DR, Diekema DJ, et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS One. 2014;9(7). doi:10.1371/journal.pone.0101510
96. Gutiérrez J, Morales P, González MA, Quindós G. Candida dubliniensis, a new fungal pathogen. J Basic Microbiol. 2002;42(3):207–227. doi:10.32388/ef9mql
97. Khan Z, Ahmad S, Joseph L, Chandy R. Candida dubliniensis: an appraisal of its clinical significance as a bloodstream pathogen. PLoS One. 2012;7(3):e32952. doi:10.1371/journal.pone.0032952
98. Pfaller MA, Messer SA, Gee S, et al. In vitro susceptibilities of Candida dubliniensis isolates tested against the new triazole and echinocandin antifungal agents. J Clin Microbiol. 1999;37(3):870–872. doi:10.1128/jcm.37.3.870-872.1999
99. Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2017;7:1–12. doi:10.3389/fmicb.2016.02173
100. Mølgaard-Nielsen D, Pasternak B, Hviid A. Use of oral fluconazole during pregnancy and the risk of birth defects. N Engl J Med. 2013;369(9):830–839. doi:10.1056/nejmoa1301066
101. Martin MV. Review The use of fluconazole and itraconazole in the treatment of. J Antimicrob Chemother. 1999;44:429–437. doi:10.1093/jac/44.4.429
102. Pasko MT, Piscitelli SC, Van Slooten AD. Formulary forum. Ann Pharmacother. 1989;24(9):860–867.
103. Sobel JD, Wiesenfeld HC, Martens M, et al. Maintenance fluconazole therapy for recurrent vulvovaginal candidiasis. N Engl J Med. 2004;351(9):876–883. doi:10.1056/nejmoa033114
104. Maertens JA. History of the development of azole derivatives. Clin Microbiol Infect. 2004;10(SUPPL. 1):1–10. doi:10.1111/j.1470-9465.2004.00841.x
105. Nielsen LE, Forrester JB, Girotto JE, Dassner AM, Humphries R. One size fits all? Application of susceptible-dose-dependent breakpoints to pediatric patients and laboratory reporting. J Clin Microbiol. 2020;58(1):1–8. doi:10.1128/JCM.01446-19
106. Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother. 2005;49(2):668–679. doi:10.1128/AAC.49.2.668-679.2005
107. Teo JQM, Lee SJY, Tan AL, et al. Molecular mechanisms of azole resistance in Candida bloodstream isolates. BMC Infect Dis. 2019;19(1):1–4. doi:10.1186/s12879-019-3672-5
108. Yao D, Chen J, Chen W, Li Z, Hu X. Mechanisms of azole resistance in clinical isolates of Candida glabrata from two hospitals in China. Infect Drug Resist. 2019;12:771–781. doi:10.2147/IDR.S202058
109. Pfaller MA, Boyken LB, Hollis RJ, et al. Validation of 24-hour fluconazole MIC readings versus the CLSI 48-hour broth microdilution reference method: results from a global Candida antifungal surveillance program. J Clin Microbiol. 2008;46(11):3585–3590. doi:10.1128/JCM.01391-08
110. Espinel-Ingroff A, Canton E, Peman J, Rinaldi MG, Fothergill AW. Comparison of 24-hour and 48-hour voriconazole MICs as determined by the Clinical and Laboratory Standards Institute Broth Microdilution Method (M27-A3 document) in three laboratories: results obtained with 2162 clinical isolates of Candida spp. and other. J Clin Microbiol. 2009;47(9):2766–2771. doi:10.1128/JCM.00654-09
111. Wang JM, Bennett RJ, Andersona MZ. The genome of the human pathogen Candida albicans is shaped by mutation and cryptic sexual recombination. MBio. 2018;9(5):1–16.
112. Sitterlé E, Maufrais C, Sertour N, Palayret M, d’Enfert C, Bougnoux ME. Within-host genomic diversity of Candida albicans in healthy carriers. Sci Rep. 2019;9(1):1–12. doi:10.1038/s41598-019-38768-4
113. Pryszcz LP, Németh T, Gácser A, Gabaldón T. Unexpected genomic variability in clinical and environmental strains of the pathogenic yeast Candida parapsilosis. Genome Biol Evol. 2013;5(12):2382–2392. doi:10.1093/gbe/evt185
114. Roman EA, Faraj SE, Gallo M, Salvay AG, Ferreiro DU, Santos J. Protein stability and dynamics modulation: the case of human frataxin. PLoS One. 2012;7(9):e45743. doi:10.1371/journal.pone.0045743
115. Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 2010;39(1):23–42. doi:10.1146/annurev.biophys.093008.131258
116. Laskowski RA, Gerick F, Thornton JM. The structural basis of allosteric regulation in proteins. FEBS Lett. 2009;583(11):1692–1698. doi:10.1016/j.febslet.2009.03.019
117. Mane A, Vidhate P, Kusro C, et al. Molecular mechanisms associated with Fluconazole resistance in clinical Candida albicans isolates from India. Mycoses. 2016;59(2):93–100. doi:10.1111/myc.12439
118. Morschhäuser J. The development of fluconazole resistance in Candida albicans – an example of microevolution of a fungal pathogen. J Microbiol. 2016;54(3):192–201. doi:10.1007/s12275-016-5628-4
119. Prakash SMU, Nazeer Y, Jayanthi S, Kabir MA. Computational insights into fluconazole resistance by the suspected mutations in lanosterol 14α-demethylase (Erg11p) of Candida albicans. Mol Biol Res Commun. 2020;9(4):155–167. doi:10.22099/mbrc.2020.36298.1476
120. Morio F, Loge C, Besse B, Hennequin C, Le Pape P. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis. 2010;66(4):373–384. doi:10.1016/j.diagmicrobio.2009.11.006
121. Lepesheva GI, Waterman MR. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all Biological Kingdoms. Biochim Biophys Acta. 2008;1770(3):467–477. doi:10.1016/j.bbagen.2006.07.018
122. Lepesheva GI, Waterman MR. Structural basis for conservation in the CYP51 family. Biochimica et Biophysica Acta. 2011;1814(1):88–93. doi:10.1016/j.bbapap.2010.06.006
123. Lepesheva GI, Waterman MR. CYP51 - the omnipotent P450. Mol Cell Endocrinol. 2004;215(1–2):165–170. doi:10.1016/j.mce.2003.11.016
124. Lepesheva GI, Virus C, Waterman MR. Conservation in the CYP51 family. Role of the B′ helix/BC loop and helices F and G in enzymatic function. Biochemistry. 2003;42(30):9091–9101. doi:10.1021/bi034663f
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.