Back to Journals » Drug Design, Development and Therapy » Volume 15
A Network Pharmacology Study on the Molecular Mechanism of Protocatechualdehyde in the Treatment of Diabetic Cataract
Authors Cheng X, Song Z, Wang X, Xu S, Dong L, Bai J, Li G, Zhang C
Received 18 August 2021
Accepted for publication 17 September 2021
Published 23 September 2021 Volume 2021:15 Pages 4011—4023
DOI https://doi.org/10.2147/DDDT.S334693
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Xiao Cheng, Zhihui Song, Xin Wang, Shanshan Xu, Liming Dong, Jie Bai, Guangyao Li, Chao Zhang
Department of Pharmacy, Beijing Tongren Hospital, Capital Medical University, Beijing, People’s Republic of China
Correspondence: Chao Zhang
Department of Pharmacy, Beijing Tongren Hospital, Capital Medical University, Beijing, People’s Republic of China
Tel/Fax +86-10-58267167
Email [email protected]
Purpose: Protocatechualdehyde (PCA) is a phenolic compound found in the roots of Salvia miltiorrhiza with anti-proliferative and antioxidant activities. At present, there are few studies on protocatechualdehyde against diabetic cataract (DC), and there is also lack of systematic research on the mechanism of protocatechualdehyde. Therefore, this study tried to comprehensively clarify the targets and complex mechanisms of PCA against DC from the perspective of network pharmacology.
Materials and Methods: Through collecting relevant targets from the databases, GO and KEGG enrichment analysis were performed on the potential targets. Moreover, core genes were identified by topological analysis of protein–protein interaction (PPI) network and gene–phenotype correlation analysis.
Results: The results indicated that protocatechualdehyde may be closely related to targets such as AKT1, MAPK3 and HDAC3, as well as signal pathways such as MAPK signaling pathway, PI3K-Akt signaling pathway and AGE-RAGE signaling pathway in diabetic complications.
Conclusion: Together, the present study systematically clarified the possible mechanisms of protocatechualdehyde in the treatment of diabetic cataract and provided new ideas for the drug research of this disease.
Keywords: protocatechualdehyde, diabetic cataract, network pharmacology, topological analysis, gene–phenotype correlation analysis
Introduction
Diabetic cataract (DC) is a common diabetic eye complication with an increasing incidence year by year. Cataract is one of the main causes of vision impairment in diabetic patients.1 According to reports, diabetic patients are 5 times more likely to develop cataract, especially in the early stages.2 Surgical removal of cataract is currently an effective treatment method. However, surgical treatment has many complications and high risks, which seriously affect the daily life and work of patients.2 Therefore, the search for drugs to prevent and treat DC has become a current research hotspot.
Protocatechualdehyde (PCA) is a phenolic compound found in the roots of Salvia miltiorrhiza with anti-proliferative and antioxidant activities.3,4 According to reports, protocatechualdehyde has strong protein glycosylation inhibitory activity,3,4 and has an inhibitory effect on the formation of AGEs-BSA in vivo and in vitro, and can inhibit the expression of AGEs and TGF-β1 receptors in human lens epithelial cells cultured under diabetic conditions.5 In streptozotocin (STZ)-induced diabetic rats, oral PCA for 8 weeks could significantly improve the development of diabetic cataract,5 which indicated that PCA may be an effective drug for the treatment of diabetic complications.
Due to the rapid development of bioinformatics, network pharmacology has become an emerging method that can effectively and systematically elucidate the molecular and pharmacological mechanisms of drugs.6,7 The concept of network pharmacology was clearly proposed by Hopkins in 2008,8 and it is defined as a branch of pharmacology that uses network methods to analyze the “multi-component-multi-target-multi-pathway” relationship between drugs, targets and diseases.6 In network pharmacology, a healthy body is maintained by a complex dynamic biological network, and the imbalance of the dynamic network will lead to the occurrence of diseases, that is, some targets or pathways in the network are in a state of functional imbalance.9 Therefore, when using network pharmacology to study the mechanism of a certain drug in the treatment of a certain disease, it is necessary to take the target in the biological network as the entry point, and analyze the association and dynamic changes between the molecules in the drug and the functional imbalance targets in the network from the perspective of the dynamic stability of the overall network.9
According to the above research, it can be seen that protocatechualdehyde improves diabetic cataract through different targets and pathways. However, there is little relevant network analysis and systematic research for these therapeutic targets and pathways. To elucidate the effects of PCA on diabetic cataract, the study applied network pharmacology method to explore the closely related targets of PCA in the treatment of diabetic cataract.
Materials and Methods
The flow chart of this study is depicted in Figure 1. First, potential targets of PCA against diabetic cataract were found through data mining based on multiple databases. Then, the OmicShare tool (http://www.omicshare.com/tools) was used to perform GO and KEGG pathway enrichment analysis on these potential targets,10 and the Metascape tool (www.metascape.org/) was used to perform module-based network analysis on these targets.11 Next, for these potential targets, the Cytoscape software (http://cytoscape.org/) was used for topological analysis of protein–protein interaction (PPI) network,12 and the VarElect online tool (http://ve.genecards.org) was used for gene–phenotype correlation analysis.13 The intersection targets of the above two analyses were likely to be the core targets of PCA against diabetic cataract. In addition, the key targets of PCA against diabetic cataract were verified by in vitro assays.
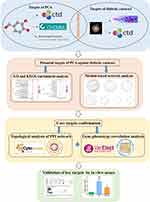 |
Figure 1 A flow chart of the entire study. |
PCA-Related Target Collection
PubChem (https://pubchem.ncbi.nlm.nih.gov/),14 as a public resource library, can provide structural information about chemical substances and their biological activities. Through the PubChem database, 3D molecular structure of PCA can be obtained. Then, based on structure similarity and the ligand-based pharmacophore model, PCA-related targets can be predicted from the ChEMBL database (http://www.ebi.ac.uk/chembl/),15 the Comparative Toxicogenomics Database (http://ctdbase.org/), SwissTargetPrediction (http://www.swisstargetprediction.ch).16 After duplicate data were discarded, acquired targets were mapped to the UniProt database (http://www.uniprot.org/) for standardization (Table S1).
Diabetic Cataract-Related Target Collection
DisGeNET (http://www.disgenet.org/) and the Comparative Toxicogenomics Database (http://ctdbase.org/) were accessed to obtain diabetic cataract-related targets using the search term “ diabetic cataract” on the platform.17 Duplicate targets were discarded and detailed information about the selected genes were described in the Supplementary Table S2.
PPI Network Construction
The intersection of PCA-related targets and diabetic cataract-related targets were considered to potential therapeutic targets of PCA against diabetic cataract. String database (https://string-db.org/, version 10.5) is a database for studying known and predicted protein–protein interactions (PPI).18 From here, the interaction of the intersection targets of PCA against diabetic cataract can be obtained, and the type is limited to “human”. It uses a confidence range to define PPI (low confidence: score <0.4; moderate: 0.4 to 0.7; height:> 0.7). Based on these scores, this study retained a PPI with a comprehensive score > 0.9. Cytoscape (http://cytoscape.org/, version 3.5.1) is a tool for visualizing complex relationships.12 Based on the PPI from String database, Cytoscape software can be used to establish a network of relationships between targets.
GO and KEGG Enrichment Analysis
The important biological processes involved in the targets can be obtained by enrichment analysis, that is, the OmicShare database can be used for GO and KEGG enrichment analysis. OmicShare tool (http://www.omicshare.com/tools) is a comprehensive set of functional annotation tool used to clarify the biological meaning of large gene data sets.10 It is often used for enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The enriched GO entries and pathways are defined as False Discovery Rate (FDR)<0.01. According to the biological process regulated by the targets, the mechanism of PCA against diabetic cataract can be analyzed.
Module-Based Network Analysis
A module-based network analysis can be conducted through Metascape (www.metascape.org/),19 which is a powerful gene function annotation analysis tool. It uses the mature MCODE algorithm to find densely connected protein groups in the network of targets and annotates the biological functions of each group.
Topological Analysis of PPI Network
Cytoscape can be used to calculate the topological parameters of all nodes in the above PPI network. For each node in the interactive network, three indices can be calculated to evaluate its topological characteristics. “Degree” is defined as the number of edges of the node i; “Betweenness Centrality” means the number of shortest paths between the pair of nodes through the node run; “Closeness Centrality” is the reciprocal of the sum of the distances from the node i to other nodes. Generally, the highest-ranking nodes based on the parameters of degree, betweenness centrality, and closeness centrality can be confirmed as the core targets.
Gene–Phenotype Correlation Analysis
The VarElect tool (http://ve.genecards.org) is a free web-based phenotype-dependent variant/gene prioritizer.13 VarElect employs GeneCards’ powerful search and scoring functions, and its algorithm provides inferred direct as well as indirect links between uploaded genes and inputted disease/phenotype. Therefore, the online tool affords a robust facility for ranking genes and pointing out their likelihood to be related to specific diseases.13 In this research, we analysed the correlation between potential therapeutic targets of PCA against diabetic cataract and the “diabetic cataract” phenotype.
Validation of Key Targets by in vitro Assays
The SRA01/04 cell line was purchased from the Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College (Beijing, China). 25 mM high glucose (HG) was accessed to induce human lens epithelial cells SRA01/04 to establish a DC cell model. After the model was successfully established, 10 μM PCA was used for drug intervention, and the non-administered group was given an equal volume of culture medium. The experiment was divided into three groups: normal control group, 25 mM high glucose treatment group, and 25 mM high glucose + 10 μM PCA group. After drug intervention for 24 hours, discard the supernatant, add 1mL TRIzol to each petri dish. The procedure of RNA extraction was referenced previously.20 The primers included AKT1, MAPK3, HDAC3 and β-actin. Primer sequences were shown in Table 1. The RNA was reversely transcribed into cDNA with PrimeScript™ RT reagent Kit. Then 2 μL cDNA templates were subjected to real-time PCR using SYBR® Premix Ex Taq II. According to the relative quantification of 2–ΔΔCt method, the expressions of target genes of AKT1, MAPK3 and HDAC3 could be determined when using β-actin as internal reference.
 |
Table 1 Primer Sequences of AKT1, MAPK3 and HDAC3 |
Results
PPI of Potential Targets of PCA Against Diabetic Cataract
A total of 366 PCA-related targets were obtained by searching the ChEMBL database, the Comparative Toxicogenomics Database, and SwissTargetPrediction. In addition, a total of 1363 diabetic cataract-related targets were acquired from DisGeNET and the Comparative Toxicogenomics Database. After duplicate data were discarded, the intersection genes were 344 potential targets of PCA against diabetic cataract. According to the confidence range of defining PPI in String database, 174 targets were reserved with a confidence score > 0.9 out of all the intersection targets. Based on the PPI from String database, Cytoscape software can be used to establish a network of relationships among targets. After removing nodes with few edges in the PPI network, 105 targets were finally retained for the following study (Figure 2). The detailed topological parameters of 105 targets were shown in the Supplementary Table S3.
 |
Figure 2 PPI network of potential targets of PCA to treat diabetic cataract. |
GO and KEGG Enrichment Analysis of Potential Targets
In order to clarify the multiple mechanisms of PCA on diabetic cataract from the system level, we used OmicShare tool to perform GO and KEGG enrichment analysis on 105 targets. The following figure listed the top 20 biological process GO entries (p <0.05), molecular function GO entries (p <0.05), cellular components GO entries (p <0.05), and KEGG signaling pathways in which these targets were involved (Figure 3).
The results showed that the potential targets of PCA against diabetic cataract were closely related to the following cell components: protein-containing complex, cytoplasm and receptor complex; closely related to the following molecular functions: protein kinase activity, protein kinase binding and protein serine/threonine kinase activity; closely related to the following biological processes: positive regulation of signal transduction, positive regulation of intracellular signal transduction, positive regulation of cell communication and positive regulation of cellular metabolic process. KEGG pathways mainly involved MAPK signaling pathway, PI3K-Akt signaling pathway and AGE-RAGE signaling pathway in diabetic complications. These results suggested that PCA may act by participating in the above-mentioned biological processes, molecular functions, cellular components and signal pathways.
Module-Based Network Analysis of Potential Targets
A module-based network analysis was conducted through Metascape. The mature MCODE algorithm was applied to find densely connected protein groups in the network of potential targets and annotate the biological functions of each group. Seven functional modules were extracted from the above 105 targets (Figure 4). Pathway and process enrichment analysis were performed on the obtained modules, and the enriched function annotations included “Diseases of signal transduction by growth factor receptors and second messengers”, “positive regulation of cell death”, and “Signaling by Interleukins”.
Topological Analysis of PPI Network
Cytoscape was used to calculate the topological characteristics of all nodes in the above-mentioned PPI network. Generally, the highest-ranking nodes based on the topological parameters of degree, betweenness centrality, and closeness centrality were confirmed as the hub targets. In this study, there were 14 possible hub genes. As shown in Figure 5 and Table 2, hub genes in the network included AKT1, APP, CASP3, CASP8, CREBBP, EP300, ESR1, HDAC1, HDAC3, HSP90AA1, MAPK1, MAPK3, PIK3CA, and SMAD3.
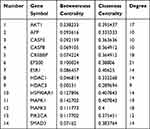 |
Table 2 Topological Parameters of Hub Genes in the PPI Network |
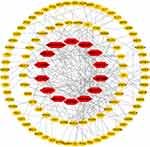 |
Figure 5 Topological analysis of PPI network. Red nodes on the inner circle indicate hub genes. |
Gene–Phenotype Correlation Analysis of Anti-DC Targets of PCA
VarElect’s algorithm provides direct as well as indirect links between genes and disease/phenotype. After analyzing the correlation between potential therapeutic targets and the diabetic cataract phenotype, the direct (orange nodes in the center) and indirect genes (blue nodes in the periphery) associated with the diabetic cataract phenotype were shown in the following figure (Figure 6). The results showed that among the 105 potential targets, 78 were directly associated and 27 were indirectly associated with the DC phenotype (Figure 6). The darker the color of the orange nodes, the larger the area of the blue nodes, indicated the closer the correlation with the disease phenotype. Besides, the top ten directly and indirectly associated genes were shown in Table 3. The higher the Average Disease Causing Likelihood, the closer the relationship between the target and DC disease.
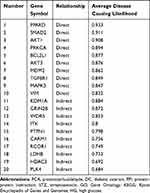 |
Table 3 Top Ten Targets Directly or Indirectly Associated with the DC Phenotype. The Average Disease-Causing Likelihood Indicates the Closeness Between Genes and the Phenotype |
Validation of Key Targets by in vitro Assays
Validation of key targets was carried out by in vitro assays. 25 mM high glucose was used to induce SRA01/04 cell to establish a DC cell model. Compared with the control group, the transcriptional levels of AKT1, MAPK3 and HDAC3 in HG-induced human lens epithelial cells were significantly increased (all P<0.01, Figure 7). However, PCA treatment significantly inhibited mRNA levels of AKT1, MAPK3 and HDAC3 in HG- induced cells (all P<0.01, Figure 7).
Discussion
Clinically, diabetic cataract occurs earlier than senile cataract, and the course of the disease progresses faster. Therefore, it is of great significance to find effective drugs to inhibit or delay diabetic cataract. However, the pathogenesis of DC is very complex, including the activation of aldose reductase (AR) receptors, the accumulation of sorbitol, galactitol and advanced glycation end products (AGEs), and oxidative stress damage activated by reactive oxygen species (ROS).21–24 At present, the effective treatment for various types of cataracts is still surgery, and there is still no medicine that can effectively inhibit or delay the occurrence and development of DC. The reason is that the purification methods of the drugs studied in these experiments are not uniform, or the way of action of drugs on diabetic cataract is relatively single, so it may be weak for the drugs on the effect of diabetic cataract with complex pathogenesis in actual clinical conditions. PCA is a phenolic compound with anti-proliferative activity, antioxidant activity, protein glycosylation inhibitory activity,3,4 and can prevent the expression of AGEs and TGF-β1 receptors in human lens epithelial cells cultured under diabetic conditions.5 According to the above studies, it can be found that PCA can improve disease progression through multiple pathways and targets, which is exactly a characteristic of therapeutic drugs for complex diseases.
Network pharmacology is a systematic research method that integrates systems biology, omics and computational biology, and is based on the interaction network between components, diseases, and targets. It can systematically reveal the mechanism of action of drugs to treat diseases.25 PCA has the characteristics of multi-target and multi-pathway effects on diseases. Compared with traditional test methods, network pharmacology can comprehensively explain the potential mechanism of action of drugs using scientific analysis methods.26 Based on the method of network pharmacology, this study analyzed the possible targets of PCA in the treatment of DC, and conducted in vitro verification experiment to improve the reliability of the results.
To evaluate the effects of PCA on diabetic cataract treatment, the network pharmacology analysis was carried out. First, a total of 105 potential targets of PCA against diabetic cataract were obtained from PPI network by searching for different databases. After that, in order to clarify the multiple mechanisms of PCA on diabetic cataract, GO and KEGG enrichment analysis were performed on these targets. The GO results showed that the potential targets of PCA against diabetic cataract were closely related to the following molecular functions: protein kinase activity, protein kinase binding and protein serine/threonine kinase activity; closely related to the following biological processes: positive regulation of signal transduction, positive regulation of intracellular signal transduction, and positive regulation of cellular metabolic process. The above results indicated that many of these targets may be protein kinases or protein serine/threonine kinases, and they may play a role in signal transduction and participation in metabolic processes, which is consistent with the results of the core targets screened out later. Moreover, the results of KEGG enrichment analysis revealed that signal pathways involved in these targets mainly include MAPK signaling pathway, PI3K-Akt signaling pathway and AGE-RAGE signaling pathway in diabetic complications. Gong et al proposed that long-chain non-coding RNA in diabetic cataract promotes the apoptosis and oxidative stress of lens epithelial cells through the p38/MAPK signaling pathway.27 TGFb/PI3K/Akt signalling in the DC pathological process could invoke the onset and progression of EMT.28 AGEs are glycosylation products formed by non-enzymatic reactions with the amino groups of large biomolecules under the stimulation of chronic hyperglycemia.29 Recent studies have confirmed that AGEs play an important pathogenic role in the development of diabetic cataract.30,31 In addition to depositing in the extracellular matrix, AGEs also bind to the AGEs receptor (RAGE) to activate NADPH oxidase (Nox) and NF-κB,32,33 thus starting the vicious circle of oxidative stress and inflammation.34,35 Since the formation of AGEs cannot be reversed, even if hyperglycemia is improved, their accumulation in the lens will continue to cause oxidative stress.36 Based on the above results, we found that the therapeutic effects of PCA on diabetic cataract not only involve multiple targets, but also multiple pathways and mechanisms. Besides, according to other reports, there were many protein kinases involved in the MAPK signaling pathway and PI3K-Akt signaling pathway.37 This suggested that, consistent with the results of GO enrichment, KEGG pathway enrichment analysis also shows that many potential targets are probably protein kinases, and they play an important role in the treatment of DC.
Next, in order to confirm the core targets of PCA against diabetic cataract, we performed topological analysis of PPI network and gene–phenotype correlation analysis of anti-DC targets of PCA. The intersection targets of the above two analyses were very likely to be the core targets of PCA against diabetic cataract. They were AKT1, MAPK3, and HDAC3. Same as the GO enrichment results mentioned above, AKT1 and MAPK3 were both protein kinases, and AKT1 belonged to the PI3K-Akt signaling pathway in the KEGG enrichment result, and MAPK3 belonged to the MAPK signaling pathway in the KEGG enrichment result. AKT1 plays an important role in regulating cell survival, and can be activated by PI3K in diabetic cataract [28]. MAPK, mitogen-activated protein kinase, is an important substance that transmits signals from the cell surface to the inside of the nucleus, participates in the regulation of cell proliferation and differentiation, and plays a key role in cell proliferation-related signal pathways.37 HDAC3 (Histone deacetylase 3) is a type of protease, which plays an important role in chromosome structural modification and gene expression regulation. HDAC is activated in age-related cataract, and the formation of cataract is alleviated by HDAC inhibition.38–40 It is possible that ocular HDAC increases in ocular diabetic complications.41 These 3 core targets exerted their anti-DC disease effects through different mechanisms, which pointed that the therapeutic effects of PCA may be multifaceted.
Finally, verification of key targets was carried out by in vitro assays. As shown in Figure 7, the transcriptional levels of AKT1, MAPK3 and HDAC3 in HG- induced human lens epithelial cells were significantly increased, and PCA treatment significantly inhibited mRNA levels of AKT1, MAPK3 and HDAC3 in HG- induced cells. The above results demonstrated that these three targets are likely to be the key targets for PCA to treat diabetic cataract, and further experimental studies are needed in the future.
This study tried to clarify some of the key targets and mechanisms of PCA for the treatment of DC, but there were still some limitations. Although we have collected a large number of databases to obtain PCA-related targets and DC-related targets, the data was certainly incomplete and some of the latest research had not been unearthed. Secondly, for the complex network of PCA regulating DC, more experiments should be still needed to verify these targets and key pathways.
Conclusion
In general, this study revealed that PCA may exert a therapeutic effect on diabetic cataract through a variety of targets and signaling pathways. Although further research is needed to clarify the exact mechanism, the current research provides a systematic and comprehensive overview of possible targets and signaling pathways related to PCA against DC.
Acknowledgments
This research was supported by the priming scientific research foundation for the senior researcher in Beijing Tongren Hospital, Capital Medical University, and the priming scientific research foundation for the junior researcher in Beijing Tongren Hospital, Capital Medical University (2020-YJJ-ZZL-049).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Drinkwater JJ, Davis WA, Davis TME. A systematic review of risk factors for cataract in type 2 diabetes. Diabetes Metab Res Rev. 2019;35(1):e3073. doi:10.1002/dmrr.3073
2. Kiziltoprak H, Tekin K, Inanc M, Goker YS. Cataract in diabetes mellitus. World J Diabetes. 2019;10(3):140–153. doi:10.4239/wjd.v10.i3.140
3. Anwar S, Khan S, Almatroudi A, et al. A review on mechanism of inhibition of advanced glycation end products formation by plant derived polyphenolic compounds. Mol Biol Rep. 2021;48(1):787–805. doi:10.1007/s11033-020-06084-0
4. Wang J, Xu J, Gong X, Yang M, Zhang C, Li M. Biosynthesis Chemistry, and pharmacology of polyphenols from Chinese salvia species: a review. Molecules. 2019;24(1):155. doi:10.3390/molecules24010155
5. Kim YS, Kim NH, Lee SW, Lee YM, Jang DS, Kim JS. Effect of protocatechualdehyde on receptor for advanced glycation end products and TGF-beta1 expression in human lens epithelial cells cultured under diabetic conditions and on lens opacity in streptozotocin-diabetic rats. Eur J Pharmacol. 2007;569(3):171–179. doi:10.1016/j.ejphar.2007.05.054
6. Lin Y, Shen C, Wang F, Fang Z, Shen G. Network pharmacology and molecular docking study on the potential mechanism of Yi-Qi-Huo-Xue-Tong-Luo formula in treating diabetic peripheral neuropathy. J Diabetes Res. 2021;2021:9941791. doi:10.1155/2021/9941791
7. Zeng L, Yang K. Exploring the pharmacological mechanism of Yanghe decoction on HER2-positive breast cancer by a network pharmacology approach. J Ethnopharmacol. 2017;6(199):68–85. doi:10.1016/j.jep.2017.01.045
8. Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25(10):1110–1111. doi:10.1038/nbt1007-1110
9. Tang F, Tang Q, Tian Y, Fan Q, Huang Y, Tan X. Network pharmacology-based prediction of the active ingredients and potential targets of Mahuang Fuzi Xixin decoction for application to allergic rhinitis. J Ethnopharmacol. 2015;24(176):402–412. doi:10.1016/j.jep.2015.10.040
10. Yang M, Chen J, Xu L, et al. A network pharmacology approach to uncover the molecular mechanisms of herbal formula Ban-Xia-Xie-Xin-Tang. Evid Based Complement Alternat Med. 2018;2018:4050714. doi:10.1155/2018/4050714
11. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
12. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
13. Stelzer G, Plaschkes I, Oz-Levi D, et al. VarElect: the phenotype-based variation prioritizer of the GeneCards suite. BMC Genomics. 2016;17(Suppl2):444. doi:10.1186/s12864-016-2722-2
14. Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–13. doi:10.1093/nar/gkv951
15. Gaulton A, Hersey A, Nowotka M, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945–D954. doi:10.1093/nar/gkw1074
16. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
17. Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford). 2015;2015:bav028. doi:10.1093/database/bav028
18. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
19. Chen Y, Yuan T, Chen D, et al. Systematic analysis of molecular mechanism of resveratrol for treating pulmonary hypertension based on network pharmacology technology. Eur J Pharmacol. 2020;5(888):173466. doi:10.1016/j.ejphar.2020.173466
20. Cheng X, Yang YL, Yang H, Wang YH, Du GH. Kaempferol alleviates LPS-induced neuroinflammation and BBB dysfunction in mice via inhibiting HMGB1 release and down-regulating TLR4/MyD88 pathway. Int Immunopharmacol. 2018;56:29–35. doi:10.1016/j.intimp.2018.01.002
21. Wang F, Ma J, Han F, et al. DL-3-n-butylphthalide delays the onset and progression of diabetic cataract by inhibiting oxidative stress in rat diabetic model. Sci Rep. 2016;13(6):19396. doi:10.1038/srep19396
22. Yu X, Zheng H, Chan MT, Wu WKK. MicroRNAs: new players in cataract. Am J Transl Res. 2017;9(9):3896–3903.
23. Armao D, Bouldin TW, Bailey RM, Hooper JE, Bharucha DX, Gray SJ. Advancing the pathologic phenotype of giant axonal neuropathy: early involvement of the ocular lens. Orphanet J Rare Dis. 2019;14(1):27. doi:10.1186/s13023-018-0957-5
24. Liu X, Gong Q, Yang L, Liu M, Niu L, Wang L. microRNA-199a-5p regulates epithelial-to-mesenchymal transition in diabetic cataract by targeting SP1 gene. Mol Med. 2020;26(1):122. doi:10.1186/s10020-020-00250-7
25. Duan XC, Huang S, Peng DY, et al. Application of network pharmacology in the study of traditional Chinese medicine formula. Chin Pharmacol Bull. 2020;36(3):303–308.
26. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi:10.1038/nchembio.118
27. Gong W, Zhu G, Li J, Yang X. LncRNA MALAT1 promotes the apoptosis and oxidative stress of human lens epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes Res Clin Pract. 2018;144:314–321. doi:10.1016/j.diabres.2018.06.020
28. Du L, Hao M, Li C, et al. Quercetin inhibited epithelial mesenchymal transition in diabetic rats, high-glucose-cultured lens, and SRA01/04 cells through transforming growth factor-β2/phosphoinositide 3-kinase/Akt pathway. Mol Cell Endocrinol. 2017;452:44–56. doi:10.1016/j.mce.2017.05.011
29. Dehnad A, Fan W, Jiang JX, et al. AGER1 downregulation associates with fibrosis in nonalcoholic steatohepatitis and type 2 diabetes. J Clin Invest. 2020;130(8):4320–4330.
30. Ruiz HH, Ramasamy R, Schmidt AM. Advanced glycation end products: building on the concept of the “common soil” in metabolic disease. Endocrinology. 2020;161(1):bqz006. doi:10.1210/endocr/bqz006
31. Maruf AA, Lip H, Wong H, O’Brien PJ. Protective effects of ferulic acid and related polyphenols against glyoxal- or methylglyoxal-induced cytotoxicity and oxidative stress in isolated rat hepatocytes. Chem Biol Interact. 2015;234:96–104. doi:10.1016/j.cbi.2014.11.007
32. Pang L, Lian X, Liu H, et al. Understanding diabetic neuropathy: focus on oxidative stress. Oxid Med Cell Longev. 2020;2020:9524635. doi:10.1155/2020/9524635
33. Chaudhuri J, Bains Y, Guha S, et al. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metab. 2018;28(3):337–352. doi:10.1016/j.cmet.2018.08.014
34. Wang Z, Zhang J, Chen L, Li J, Zhang H, Guo X. Glycine suppresses AGE/RAGE signaling pathway and subsequent oxidative stress by restoring Glo1 function in the aorta of diabetic rats and in HUVECs. Oxid Med Cell Longev. 2019;2019:4628962.
35. Kopytek M, Ząbczyk M, Mazur P, Undas A, Natorska J. Accumulation of advanced glycation end products (AGEs) is associated with the severity of aortic stenosis in patients with concomitant type 2 diabetes. Cardiovasc Diabetol. 2020;19(1):92. doi:10.1186/s12933-020-01068-7
36. Bheda P. Metabolic transcriptional memory. Mol Metab. 2020;38:100955. doi:10.1016/j.molmet.2020.01.019
37. Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118(Pt 16):3569–3572. doi:10.1242/jcs.02470
38. Wang Y, Li F, Zhang G, Kang L, Guan H. Ultraviolet-B induces ERCC6 repression in lens epithelium cells of age-related nuclear cataract through coordinated DNA hypermethylation and histone deacetylation. Clin Epigenetics. 2016;26(8):62. doi:10.1186/s13148-016-0229-y
39. Xie L, Santhoshkumar P, Reneker LW, Sharma KK. Histone deacetylase inhibitors trichostatin A and vorinostat inhibit TGFβ2-induced lens epithelial-to-mesenchymal cell transition. Invest Ophthalmol Vis Sci. 2014;55(8):4731–4740. doi:10.1167/iovs.14-14109
40. Chen X, Xiao W, Chen W, Luo L, Ye S, Liu Y. The epigenetic modifier trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition of lens epithelial cells. Cell Death Dis. 2013;4(10):e884. doi:10.1038/cddis.2013.416
41. Kanada F, Takamura Y, Miyake S, et al. Histone acetyltransferase and Polo-like kinase 3 inhibitors prevent rat galactose-induced cataract. Sci Rep. 2019;9(1):20085. doi:10.1038/s41598-019-56414-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




