Back to Journals » International Journal of Nanomedicine » Volume 18
A Naïve Phage Display Library-Derived Nanobody Neutralizes SARS-CoV-2 and Three Variants of Concern
Authors Wu D, Cong J, Wei J, Hu J, Sun W, Ran W, Liao C, Zheng H, Ye L
Received 28 June 2023
Accepted for publication 7 October 2023
Published 16 October 2023 Volume 2023:18 Pages 5781—5795
DOI https://doi.org/10.2147/IJN.S427990
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
Dandan Wu,1,* Junxiao Cong,1,* Jiali Wei,1 Jing Hu,1 Wenhao Sun,1 Wei Ran,2 Chenghui Liao,1 Housheng Zheng,1 Liang Ye1
1Department of Immunology, International Cancer Center, Shenzhen University Health Science Center, Shenzhen, People’s Republic of China; 2State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, the First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liang Ye, Department of Immunology, International Cancer Center, Shenzhen University Health Science Center, Shenzhen, 518055, People’s Republic of China, Tel +86-0755-26631420, Fax +86-0755-86671943, Email [email protected]
Background: The emergence of the coronavirus disease 2019 (COVID-19) pandemic and the new severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants of concern (VOCs) requires the continuous development of safe, effective, and affordable prevention and therapeutics. Nanobodies have demonstrated antiviral activity against a variety of viruses, providing a new candidate for the prevention and treatment of SARS-CoV-2 and its variants.
Methods: SARS-CoV-2 glycoprotein spike 1 subunit (S1) was selected as the target antigen for nanobody screening of a naïve phage display library. We obtained a nanobody, named Nb-H6, and then determined its affinity, inhibition, and stability by ELISA, Competitive ELISA, and Biolayer Interferometry (BLI). Infection assays of authentic and pseudotyped SARS-CoV-2 were performed to evaluate the neutralization of Nb-H6. The structure and mechanism of action were investigated by AlphaFold, docking, and residue mutation assays.
Results: We isolated and characterized a nanobody, Nb-H6, which exhibits a broad affinity for S1 and the receptor binding domain (RBD) of SARS-CoV-2, or Alpha (B.1.1.7), Delta (B.1.617.2), Lambda (C.37), and Omicron (BA.2 and BA.5), and blocks receptor angiotensin-converting enzyme 2 (ACE2) binding. Moreover, Nb-H6 can retain its binding capability after pH or thermal treatment and effectively neutralize both pseudotyped and authentic SARS-CoV-2, as well as VOC Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (BA.2 and BA.5) pseudoviruses. We also confirmed that Nb-H6 binds two distinct amino acid residues of the RBD, preventing SARS-CoV-2 from interacting with the host receptor.
Conclusion: Our study highlights a novel nanobody, Nb-H6, that may be useful therapeutically in SARS-CoV-2 and VOC outbreaks and pandemics. These findings also provide a molecular foundation for further studies into how nanobodies neutralize SARS-CoV-2 and variants and imply potential therapeutic targets for the treatment of COVID-19.
Keywords: COVID-19, antiviral medication, single-domain antibody, neutralizing antibody, bio-screening
Introduction
The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) resulted in a global human health and economic crisis, having exceeded 763 million cases and 6.9 million deaths worldwide as of April 2023 (https://www.who.int/, accessed April 16, 2023). Despite the development of effective vaccines, eradicating SARS-CoV-2 remains extremely difficult due to waning immune responses from vaccination and the spread of new variants of concern (VOCs) such as Alpha, Beta, Gamma, and Delta, as well as the recently identified Omicron.1–4 Given the ongoing SARS-CoV-2 pandemic and the possibility of new variants continuing to emerge, efforts to develop effective prophylactic and therapeutic strategies against emerging VOCs are urgently required.
The SARS-CoV-2 glycoprotein spike (S) is a homotrimeric transmembrane protein that is responsible for viral entry into the host cell.5–7 The S protein is composed of two functional subunits, S1 and S2, which are required for binding host cell angiotensin-converting enzyme 2 (ACE2) and catalyzing virus-cell membrane fusion.5,7–9 The S1 subunit is further divided into a C-terminal receptor-binding domain (RBD) and an N-terminal domain (NTD), while the RBD directly binds to ACE2 to initiate cell recognition.5,6 To date, RBD and NTD have been identified as targets of numerous neutralizing antibodies and inhibitors,10–13 underlining that blocking S1 is crucial for the development of protective immunity against SARS-CoV-2 infection.
The single-domain antibody (also known as nanobody, VHH, or Nb) derived from the heavy chain antibody of camelids has shown many advantages over conventional antibodies.14 As one-tenth of the size of conventional antibodies, nanobodies have unique properties such as high binding activity and specificity for a target antigen, strong tissue penetration, intrinsic stability, ease of production at a low cost, and ease of genetic engineering.14,15 It is widely applied as a promising diagnostic and therapeutic tool for human diseases, including respiratory virus infections.15–17 Patients with COVID-19 are at a high risk of developing severe disease, but it has been demonstrated that neutralizing nanobodies can effectively reduce mortality.18,19 To identify novel nanobodies that might lead to advancements in diagnostics and provide new treatment options, screening of nanobodies in the naïve phage library is the preferred approach.14 Selection from a naïve phage library allows faster production of affinity antibodies compared to immunization, which is especially appealing when developing antibodies against newly emerged and rapidly spreading diseases like COVID-19.11,20 Moreover, the naïve phage library enabled the isolation of nanobodies against a wide range of antigens, including autoantigens, non-immunogenic, or toxic antigens, while avoiding the time-consuming and costly procedure of immunization. Although previous studies have shown that monomer nanobodies derived from naive phage libraries usually lack the high binding affinity required for therapeutic applications, this can be improved by fusing the Fc domain of antibodies or linking monomer nanobodies to produce dimerization or multimerization.14,20–22 When the capacity of the naïve phage library is sufficient, high-affinity antibodies can be obtained in a short period of time. Therefore, screening nanobodies from a naïve phage library for S1 specifically might be a viable and efficient strategy for combating SARS-CoV-2 and variant infection and transmission.
In this study, we used a naïve nanobody phage library to develop an anti-SARS-CoV-2 nanobody (Nb-H6) that shows an affinity for S1 and RBD of SARS-CoV-2 and three VOCs, has high thermostability and pH stability, and effectively occludes ACE2 interaction. Moreover, we determined that Nb-H6 can broadly neutralize the authentic SARS-CoV-2 as well as Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (BA.2 and BA.5) pseudoviruses in vitro. Finally, we revealed that the mechanism of Nb-H6 neutralization is interference with RBD regions Tyr449 and Tyr489 bound to ACE2. Collectively, these results suggest that Nb-H6 may be a potential therapeutic nanobody against COVID-19 caused by SARS-CoV-2 variants.
Materials and Methods
Screening Nanobodies Against SARS-CoV-2 S1 Protein
The nanobodies (Nbs) targeting Wuhan wild-type S1 (defined as S1) (Z03485, Genscript) were selected through three consecutive phage display rounds using a previously constructed naïve library.23 The naïve library was established based on proximate 1.06×1010 PBMCs isolated from 103 alpacas and possesses a capacity of 1.36×1019 individual transformants (AlpalifeBio, China). For screening of Nb candidates, 50 μg of S1 protein (GenScript Biotech Corporation, China) was coated overnight at 4 °C on Nunc Immunotubes (444202, Thermo Fisher). Following three PBS washes and 2 hours of blocking at RT with 5% BSA (A1933, Sigma-Aldrich), 100 μL of phages were added into S1-coated tubes and incubated for 1 hour at RT. After washing with PBS containing 0.05% (v/v) Tween-20 (A100777, Sangon) (PBST) 10 times, bound phages were eluted with 0.25 mg/mL Trypsin (A003702, Sangon) for 30 min at RT. Subsequent 2-round screenings were performed as described above, with a sublibrary from each round of panning requiring amplification and titration. Each round of screening used approximately 1.2×1012 phage particles. From the third-round screening sublibrary, we randomly selected 96 phage colonies for Phage-ELISA, and the top three positive colonies targeting S1 were sequenced.
Expression and Purification of SARS-CoV-2 S1-Specific Nanobodies and Related Mutants
The DNA sequences of selected nanobodies were optimized before being inserted with polyhistidine (His) and hemagglutinin (HA) tags and cloned into pET-23a (+) plasmids (General Biol, China). The constructed plasmids were heat-shock transformed into E. coli BL21 (DE3) cells (CD601, TransGen). The cells were cultured in LB medium with 100 μg/mL ampicillin until OD600 = 0.6, then induced for 15 hours at 16 °C with 0.5 mM IPTG. Cells were precipitated from culture supernatant for isolation and purification, and whole proteins from cells were extracted using ultrasonication. The S1-specific nanobodies were then purified using the HisTrap FF column (GE17-5255-01, Cytiva). Harvested target Nbs were determined by SDS-PAGE, and the concentration was measured by the Pierce™ BCA Protein Assay Kit (23225, Thermo Fisher). The expression plasmids containing mutations in Nb-H6 (named Nb-H6 N72A and Nb-H6 R97A, respectively) were synthesized by General Biol (Chuzhou, China). The Nb-H6 N72A and Nb-H6 R97A recombinant proteins were generated using the same expression and purification procedures as described above, and the binding capability of Nb-H6 N72A and Nb-H6 R97A to S1 or RBD was assessed using an ELISA and a BLI assay.
Enzyme-Linked Immunosorbent Assay (ELISA)
The binding activity of the obtained Nbs to the S1 or RBD was assessed by ELISA. In brief, a Maxisorp 96-well immune plate (446469, Thermo Fisher) was coated overnight at 4 °C with 0.4 μg of S1 or RBD (Z03479, GenScript) antigen. The plate was blocked with 5% BSA before being washed five times with PBST. Following that, 100 μL/well of different dilutions of Nbs were incubated in the plate for 1 hour at RT. Anti-HA-tag mAb-HRP-DirecT (M180-7, MBL) was added and incubated for 1 hour at RT with slight shaking. Following a PBST wash, a 100 μL/well 3.3’,5,5’-Tetramethylbenzidine (TMB) substrate (H101-01, TransGen) was added to detect horseradish peroxidase (HRP) activity. After stopping the enzyme reaction with 50 μL of 2 M H2SO4, the absorbance at 450 nm was measured with a SpectraMax 340PC Microplate Reader (Perkin-Elmer, USA).
A competitive ELISA was conducted to determine the capacity of Nb-H6, Nb-H6 N72A, and Nb-H6 R97A to inhibit the binding of S1 or RBD to hACE2. A 96-well immuno plate was coated with recombinant hACE2 (T101301, East Mab) at 400 ng per well. Following washing and blocking, serially diluted Nb-H6, Nb-H6 N72A, or Nb-H6R97 was mixed with S1 or RBD for 1 hour before being added into the coated wells to co-incubate with hACE2 for 1 hour at RT. Afterward, anti-S1 antibody (HRP) (ABIN6953153, Antibodies) or anti-RBD antibody (HRP) (10–054, ProSci) was incubated in the wells for 1 hour at RT with slight shaking. A 100 μL TMB substrate was added, followed by 50 μL of 2 M H2SO4 to terminate the enzyme reaction, and absorbance readings at OD450 were taken with a microplate reader. A half-maximal inhibitory concentration (IC50) of Nb-H6 was calculated using four-parameter logistic regression.
Biolayer Interferometry (BLI)
To measure the affinity of Nb-H6 binding to spike proteins of SARS-CoV-2 variants (Alpha B.1.1.7, Delta B.1.617.2, Lambda C.37 (East Mab, China, 40591-V08H31 Sino Biological), Omicron BA.2 (40891-V08H43 Sino Biological, EVV00324 Antibody System), and Omicron BA.5 (40592-V08H130 Sino Biological), Biolayer Interferometry (BLI) assay was performed using the BLItzTM Label-free Protein Analysis System (ForteBio, USA) with High Precision Streptavidin (SAX) Biosensors (18–5117, Sartorius).24 To label the S1 and RBD, proteins were incubated with NHS-Biotin (C100212, Sangon) at a molar ratio of 12:1 NHS-Biotin to protein for 1 hour at 4 °C and excess NHS-Biotin molecules were removed by overnight dialysis. This step allowed for the attachment of biotin molecules onto these proteins, enabling their immobilization on the SAX biosensors before Nb-H6 detection. The SAX biosensors were balanced in PBS for 10 minutes before detection, and then the biotinylated S1 or RBD antigens were immobilized on pretreated SAX sensors for 240 seconds. After a baseline stabilization period of 120 seconds in PBS, several diluted Nbs (0.25–8 μM) in PBS were incubated with coated biosensors for another 240 seconds. The sensors were then soaked in PBS for 240 seconds to dissociate. Additionally, the binding capability of Nb-H6 mutants to S1 or RBD was assessed using the same protocol as the BLI assay. According to the raw data, kinetic curves were recorded and fitted using a 1:1 binding model. Finally, the rate constants of association (Kon), rate constants of dissociation (Koff), and equilibrium dissociation constants (KD) were determined by BLItz Pro 1.3.0.2 software.
Evaluation of Thermal and pH Stability
Nb-H6 was dissolved in PBS with a series of pH values to detect pH stability and incubated for 2 hours at RT. To evaluate thermal stability, Nb-H6 was incubated at various temperatures for 5 min and at 90 °C for different durations ranging from 5 to 105 min. The binding capacity of all treated Nb-H6 to S1 was determined by ELISA at RT after pH or thermal treatment.
Authentic SARS-CoV-2 Neutralization Assay
The original SARS-CoV-2 strain was isolated from COVID-19 patients and preserved in the Guangzhou Customs District Technology Center BSL-3 Laboratory. The live virus neutralization assay was conducted in a BSL-3 facility. Briefly, 2×104/well Vero E6 cells (HTX1534, OTWO Biotech) were seeded into a 96-well plate. Subsequently, Nb-H6 was 4-fold serially diluted and mixed with the same volume of SARS-CoV-2 (200 FFU/well) and incubated in wells at 37 °C for 1 hour. The mixture was then removed, and 100 μL of 1.6% sodium carboxymethyl cellulose (CMC) was added to each well. The plates were incubated in a CO2 incubator at 37 °C for 24 hours. Subsequently, plates were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100. To detect the neutralization titers by focus formation assay (FFA), cells were then stained with rabbit anti-SARS-CoV-2 N protein polyclonal antibody (40143-T62, Sino Biological) at 37 °C for 24 hours. Subsequently, plates were washed with PBST three times, and followed by an HRP-labeled secondary antibody (109–035-088, Jackson ImmunoResearch). The foci were visualized by TrueBlue Peroxidase Substrate (KPL, USA), and counted with an ELISPOT reader (Cellular Technology, USA).
Pseudovirus Neutralization Assay
For infection with pseudovirus, 2×104 hACE2-expressing HEK293T cells (RM02456, ABclonal) were inoculated in a cell-culture 96-well plate and cultured overnight at 37°C in DMEM (11965092, Thermo Fisher) medium with 10% fetal bovine serum (FBS, FS301- 02, TransGen) and 1 × Penicillin-Streptomycin (P/S) (FG101-01, TransGen). The SARS-CoV-2 spike pseudotyped (Wildtype/B.1.1.7/B.1.617.2/BA.2/BA.5) GFP-Luciferase lentivirus (GM-0220PV07/GM-0220PV33/GM-0220PV45/GM-0220PV86/GM-0220PV90, Genomeditech), whose final concentration was 2.75×105 TU/mL, was mixed with different dilutions of Nb-H6 for 1 hour at RT. Afterward, cells were infected with Nb-H6-pretreated SARS-CoV-2 spike pseudoviruses for 6 hours before being switched to a fresh medium. Green fluorescence images were captured 48 hours after infection using a B80iF Inverted Biofluorescence Microscope System (Daoyi, China). For determining the neutralization activity of Nb-H6, luciferase reactions of infected cells were carried out using a Single Luciferase Report Assay Kit (FR101, TransGen), and relative light units (RLUs) were detected with a SpectraMax Gemini XPS Microplate Reader (Molecular Devices, USA). Based on the obtained results, IC50 was further analyzed using four-parameter logistic regression.
Structure Modeling with AlphaFold
The structure of Nb-H6 was predicted using the AlphaFold Colab notebook with default parameters.25 (AlphaFoldv2.1.0.https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb). The input amino acid sequence of Nb-H6 will serve as a template for homology-based sequence searching. AlphaFold’s deep learning techniques were then employed to generate a 3D structural model. The notebook calculated an IDDT (Internal Distance-based Threshold Distance) score for the predicted models. The model with the highest IDDT score is typically considered the best prediction, as it indicates a higher degree of structural accuracy. The predicted model with the highest IDDT score was downloaded and visualized by PyMOL software (Schrödinger).
Docking of RBD with Nb-H6
The Nb-H6 molecular docking was carried out on the COVID-19 Docking Server (http://ncov.schanglab.org.cn/) by choosing RBD as the SARS-CoV-2 protein target. The server returned several candidate results based on the structure of Nb-H6. To choose the best candidate result, we evaluated the prediction quality based on its similarities with the published nanobody against the SARS-CoV-2 spike structure (PBD code: 7KKL),21 and its ranking in the docking score. The prediction result that ranked first and had the highest consistency score was chosen for further analysis. The interface residues at 72 (located in framework region 3 (FR3)) and 97 (located in complementarity-determining region 3 (CDR3)) of Nb-H6 and residues 449 and 489 of RBD were determined for restraint docking. The 3D structures of Nb-H6 and RBD were finally visualized by the software PyMol (Schrödinger).
Statistical Analyses
GraphPad Prism software (version 8.0) was used for statistical analysis of data and graph generation. Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA with Dunnett’s multiple comparisons test as indicated in the relative figure legends.
Results
Screening and Identification of Nanobodies Against the SARS-CoV-2 S1 Protein
High-throughput naïve nanobody libraries are more widely available to recognize multiple target antigens quickly to generate nanobodies while avoiding undesirable biochemical properties including destabilization, multi-reactivity, and agglomeration during affinity maturation.14,26 To obtain potent SARS-CoV-2 neutralizing nanobodies, we used standard phage display technology and performed three consecutive rounds of phage display, followed by a specific SARS-CoV-2 S1 protein-based binding screen using a phage ELISA (Figure 1a). As shown in Figure 1b, the phage clones specific to the S1 protein were effectively enriched over 149-fold after three rounds of panning. A total of 96 phage clones were randomly inoculated and screened for specific binding to the S1 protein by ELISA (Figure 1c). The result demonstrated that the OD450 nm value of three clones targeting S1 was six times higher than the blank control coating with BSA (Figure 1d), which was further identified by the amino acid sequences of their complementary determining regions (CDRs) (Figure 1e) and named Nb-H6, Nb-B7, and Nb-G7, respectively.
Production and Identification of Nanobodies Against SARS-CoV-2 S1 and RBD
The His and HA tag-labeled nanobodies (Nb-H6, Nb-B7, and Nb-G7) were produced by IPTG-induced E. coli BL21 (DE3) cells and further purified by the HisTrap FF column. The SDS-PAGE analysis revealed that the purified Nb-H6, Nb-B7, and Nb-G7 proteins had the expected apparent molecular weight (Figure 2a). To further confirm the specificity of binding of these three nanobodies, SARS-CoV-2 S1 or RBD antigen coated on a plate was employed to capture Nb-H6, Nb-B7, and Nb-G7. The results indicated that Nb-H6 had the greatest affinity for S1 and RBD of the three nanobodies (Figure 2b), implying that Nb-H6 could be used as an ideal nanobody for further investigation.
Nb-H6 Binds to SARS-CoV-2 S1 and RBD and Blocks ACE2 Binding
Since Nb-H6 showed the strongest binding activity, we further assessed the binding kinetics of Nb-H6 with S1 and RBD by biolayer interferometry (BLI). The results showed that Nb-H6 had an excellent affinity for both S1 (KD = 0.632 μM) and RBD (KD = 0.805 μM) (Figures 3a and c). To investigate whether Nb-H6 interferes with the binding of S1 to ACE2 or RBD to ACE2, we performed competitive binding assays by ELISA. We found that Nb-H6 was able to compete with recombinant S1 to bind ACE2 with IC50 values of 1.526 μM, and 1.294 μM of Nb-H6 was also sufficient to block 50% of ACE2 binding to RBD (Figures 3b and c). Furthermore, Nb-H6 could barely interact with the RBD of MERS-CoV, showing expected specificity on binding to SARS-CoV-2 (Supplementary Figure 1). These findings demonstrate that Nb-H6 is a tool valuable for capturing SARS-CoV-2 spikes and competitive binding studies.
Nb-H6 Shows Excellent Thermal and pH Stability
To test the biophysical stability of Nb-H6, we exposed it to a variety of denaturing conditions, including various pH values and different temperatures. As shown in Figure 4, after 5 minutes of incubation at 4, 30, 60, and 90 °C, Nb-H6 can still retain complete affinity (Figure 4a). Furthermore, the binding activity of Nb-H6 remained at 50% even after 90 minutes of incubation at 90 °C (Figure 4b). Likewise, the binding activity of Nb-H6 was not affected by low or high pH incubation for 2 hours (Figure 4c). The results presented above demonstrate that highly stable nanobodies can be rapidly and efficiently generated by the naïve phage display platform, and Nb-H6 might be beneficial for viral neutralization.
Nb-H6 Inhibits SARS-CoV-2 Infection
To verify the neutralizing capacity of Nb-H6 against SARS-CoV-2 infection, we performed in vitro neutralization assays with pseudotyped and authentic SARS-CoV-2, respectively. The pseudovirus infection results suggested that Nb-H6 could effectively block the entry of SARS-CoV-2 pseudovirus to HEK293-hACE2 cells in a dose-dependent manner (Figure 5a, left panel). By luciferase reaction analysis, the IC50 of Nb-H6 neutralizing the SARS-CoV-2 pseudovirus was determined to be 7.16 μM (Figure 5a, right panel). Accordingly, we further examined the antiviral ability of Nb-H6 against the original SARS-CoV-2 virions. As expected, Nb-H6 potently inhibited authentic SARS-CoV-2 infection in Vero E6 cells with an IC50 value of 1.975 μM (Figure 5b). These results indicate that Nb-H6 may have potential for the treatment of SARS-CoV-2 infection.
Nb-H6 Effectively Binds SARS-CoV-2 S1 and RBD Variants
Because SARS-CoV-2 new variants of concern (VOCs) strains can evade current neutralizing antibodies, we sought to investigate if Nb-H6 has a certain affinity for SARS-CoV-2 VOCs to have broad-spectrum neutralizing effects. We analyzed binding affinities of Nb-H6 toward S1 of SARS-CoV-2 VOCs (Alpha, Delta, Lambda, and Omicron) using BLI, and found that Nb-H6 showed similar affinity to S1 from B.1.1.7 (Alpha) (KD = 1.084 μM), B.1.617.2 (Delta) (KD = 1.119 μM), and C.37 (Lambda) (KD = 1.574 μM), with a slight increase in affinity to S1 from BA.2 (Omicron) (KD = 2.476 μM) (Figures 6a and c). Similarly, Nb-H6 had an affinity for the RBD of SARS-CoV-2 VOCs: Delta B.1.617.2, Lambda C.37, and Omicron BA.2 and BA.5, with KD values of 1.894 μM, 1.523 μM, 2.226 μM, and 1.700 μM, respectively (Figures 6b and c).
Nb-H6 Suppresses SARS-CoV-2 Variant Pseudovirus Infection
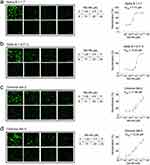
Regarding the favorable affinity of Nb-H6 binding to SARS-CoV-2 S1 and RBD variants, we sought to investigate whether Nb-H6 could also be effective against recently discovered VOCs B.1.1.7 (Alpha), B.1.617.2 (Delta), BA.2 (Omicron), and BA.5 (Omicron) in vitro. As expected, Nb-H6 inhibited the numbers of GFP-positive cells in HEK293-hACE2 cells following SARS-CoV-2 pseudovirus variants B.1.1.7 (Alpha) (Figure 7a), B.1.617.2 (Delta) (Figure 7b), BA.2 (Omicron) (Figure 7c), or BA.5 (Omicron) (Figure 7d) infection. The pseudovirus luciferase assay further confirmed the neutralization potency of Nb-H6 against all variants with IC50 values of 5.13 μM for B.1.1.7 (Figure 7a), 10.62 μM for B.1.617.2 (Figure 7b), 17.99 μM for BA.2 (Figure 7c), and 12.35 μM for BA.5 (Figure 7d), in line with the measured affinities. These results imply that Nb-H6 could be an effective SARS-CoV-2 and variant-specific neutralizing nanobody.
Nb-H6 Neutralization Mechanism Against SARS-CoV-2
To better understand the mechanism of Nb-H6-mediated broad neutralization of SARS-CoV-2 and variants, we applied the AlphaFold tool to model the structure of Nb-H6 and used a molecular docking assay to characterize the interactions between RBD and Nb-H6. The Nb-H6 residues N72 and R97, which are found in the FR3 and CDR3 regions, were shown to be responsible for forming hydrogen bonds with Y449 and Y489 of the RBD, respectively (Figure 8a). We further found that Y449 and Y489 are located within the receptor binding motif (RBM) (Figure 8b, blue box) and are highly conserved across the RBD of both SARS-CoV-2 and VOCs via amino acid sequence alignment (Figure 8b, red box), implying that Nb-H6 may have a broad-spectrum neutralizing effect against SARS-CoV-2 and VOCs. The binding properties of Nb-H6 are similar to those of class 1 nanoantibodies as previously classified.27–29 To further verify whether N72 and R97 of Nb-H6 are critical for the binding of RBD, we mutated N72 and R97 to A, thus naming the mutation nanobodies Nb-H6 N72A and Nb-H6 R97A. The enzyme-linked immunosorbent assay (ELISA) results suggested that the absence of N72 and R97 of Nb-H6 almost completely abolished the binding between Nb-H6 and the S1 and RBD (Figure 8c). In agreement with this data, Nb-H6 N72A and Nb-H6 R97A greatly decrease their binding to S1 or RBD, as verified by the BLI assay (Figure 8d). As expected, Nb-H6 N72A and Nb-H6 R97A lost their ability to inhibit S1 and RBD binding to receptor ACE2 (Figure 8e). To validate the RBD epitopes reported in docking data, Y489 and Y499 of RBD were mutated to A, respectively. The affinity of RBD mutants with Nb-H6 was determined by ELISA and BLI tests, which revealed that mutations on Y489 or Y499 weakened the binding between Nb-H6 and RBD (Figure 8f). As a result, the capability of Nb-H6 to neutralize SARS-CoV-2 can be attributed to residues Asn72 and Arg97 competitively interacting with the conservation residues Y489 and Y499 of RBD.
Discussion
The ongoing SARS-CoV-2 pandemic poses a serious threat to global public health. Additional vaccines, neutralizing antibodies, or antiviral agents are essential to overcome this challenge. Although the current COVID-19 vaccine has a certain effect on the SARS-CoV-2 infection, the emergence of VOCs and waning humoral immunity from vaccination raise the risk of SARS-CoV-2 outbreaks.1,3,4 Novel therapeutic strategies will prioritize antiviral drugs with high yield, convenient storage and transportation, and low-cost and effective defense against the SARS-CoV-2 pandemic and SARS-CoV-2 VOC transmission.
In this study, we isolated and characterized a reliable neutralizing nanobody, Nb-H6, from an alpaca naïve phage library, which has an affinity for the S1 and RBD derived from SARS-CoV-2 or three VOCs. More importantly, we demonstrated that Nb-H6 has extremely stable thermal and pH properties and widely competes with the receptor ACE2 to bind S1 and RBD of SARS-CoV-2 and VOCs, significantly restraining pseudotyped and authentic SARS-CoV-2 infection and the entry of three VOC pseudoviruses into host cells. By predicting the structure of NB-H6 and simulating its molecular interaction with RBD, we further confirmed the mechanism by which Nb-H6 neutralizes SARS-CoV-2 and VOCs.
Traditional antibodies necessitate mammalian cell expression systems, which are time-consuming. However, nanobodies can be expressed more easily and efficiently through bacteria, yeast, and fungi.14 Moreover, nanobodies have thermal and pH stability, which means they will not be limited to antibody transportation and preservation for clinical application.15 In the current study, we successfully screened nanobodies through a phage library technology. This strategy can be widely used to isolate functional nanobodies for various antigens. One of the major concerns about COVID-19 vaccines is the antibody-dependent enhancement (ADE) effect induced by the SARS-CoV-2 spike protein antibody.30–34 Studies have revealed that SARS-CoV-2 antibody-mediated ADE is frequently mediated by Fc receptors for immunoglobulin G (IgG) (FcRs), complement receptors (CRs), or both and is most commonly observed in mononuclear phagocytes and B cells.33,35,36 Furthermore, ACE2 and spike-protein conformational changes have been linked to ADE mechanisms.37,38 However, neutralizing antibodies appear to induce ADE very little compared to non-neutralizing antibodies.36,39 Since nanobody Nb-H6 lacks the Fc domain and has a short half-life like other neutralizing antibodies,34 it might be a promising option for neutralizing balance and ADE side effects for COVID-19 therapeutics.
RBD and NTD of S1 protein are both potent parts for SARS-CoV-2 binding host cells, while numerous antibodies targeting these sites showed a strong neutralization role in SARS-CoV-2 infection.10,12,18,40 These findings suggest that screening nanobody candidates that bind S1 against SARS-CoV-2 infection may be an ideal strategy. Recent studies have described the strong neutralizing activities of RBD-directed nanobodies against SARS-CoV-2,11,13,21,41–47 whereas few reports on full length-S1-directed antibodies are capable of neutralizing a broad range of SARS-CoV-2 and VOCs. Here, we obtained an attractive nanobody, Nb-H6, which has a broad affinity for SARS-CoV-2 S1 and RBD, as well as VOCs S1 and RBD, by screening for anti-SARS-CoV-2 S1 antigen. Moreover, Nb-H6 can compete with ACE2 for S1 and RBD of SARS-CoV-2 and also prove to be effective for three variants. These features further supported the potent neutralization efficacy of Nb-H6 for SARS-CoV-2 and VOCs ((Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (BA.2 and BA.5)) pseudoviruses entry, with the exception of Omicron, which was slightly resistant to Nb-H6. To face the growing number of SARS-CoV-2 variants, AlphaFold provides a quick computational approach for predicting protein structures with high accuracy.25,48 We predicted the complex of Nb-H6 binding to SARS-CoV-2 RBD and found that Asn72 and Arg97 in the Nb-H6 CDR3 can bind to RBD by forming hydrogen bonds with residues Tyr449 and Tyr489. According to the structure determination of RBD binding to ACE2, these two critical sites lie within the RBD receptor binding motif and can strongly interact with the residues of ACE2 by forming hydrogen bonds.5 These characteristics implied that Nb-H6 could directly compete with ACE2 for RBD binding. Experimental verification of these predicted data demonstrated that mutations on any of the residues forming hydrogen bonds at the interface between Nb-H6 and RBD could undermine the binding. Indeed, the RBD residues that interact with Nb-H6 are highly conserved across SARA-CoV-2 variants,49 indicating Nb-H6 may be capable of preventing SARS-CoV-2 variants from escaping. It is worth noting that Nb-H6 also exhibits excellent in vitro thermostability and pH stability, high production yields, and ideal size, all of which may help in the fight against emerging SARS-CoV-2 variants.
Although the SARS-CoV-2 and VOCs neutralizing effect of Nb-H6 was identified in vitro, in vivo evaluations are still pending. Because the nasal mucosa is the site of infection and transmission for SARS-CoV-2, restricting the virus in the upper respiratory tract is crucial to preventing viral infection and transmission and developing COVID-19.50–52 In animal models, intranasal administration of neutralizing antibodies can prevent the infection of SARS-CoV-2 and variant strains.16,53–55 Moreover, inhalable nanobody therapy for the COVID-19 model has shown promising treatment efficacy.56–58 As a result, the assessment of Nb-H6 delivered via the mucosal route for the prevention and treatment of COVID-19 and SARS-CoV-2 transmission warrants further investigation.
Conclusions
To summarize, we developed a SARS-CoV-2 S1-specific nanobody, Nb-H6, which reveals a broad affinity with S1 and RBD of SARS-CoV-2 and of three VOCs in vitro while also disrupting S1-ACE2 and RBD-ACE2 interactions. Furthermore, in vitro, Nb-H6 can effectively neutralize both pseudotyped and authentic SARS-CoV-2, as well as the Alpha, Delta, and Omicron VOC pseudoviruses, where the mechanism of action is dependent on the binding of two amino acid residues of RBD. As a result, Nb-H6 derived from a phage library could be a reliable antiviral agent for clinical settings in preventing and treating COVID-19.
Data Sharing Statement
All data are available upon request by contact with the corresponding author.
Ethics Approval
Since IRB approval is not necessary for this study, approval from an ethics committee was not required.
Acknowledgments
This study was supported by grants from the Guangdong Basic and Applied Basic Research Foundation (2021A1515010917 and 2020A1515110410), the Guangdong Medical Science and Technology Research Foundation (A2021336), the Pearl River Talent Project of Guangdong Province (2021QN02Y426), the National Nature Science Foundation of China (32170937), the Shenzhen Science and Technology Program (RCBS20200714114958310 and 20200803131335002), the Shenzhen Peacock Plan Project (827/000655), and SZU Top Ranking Project (86000000210) to Liang Ye. We sincerely thank Dr. Huawei Wei at East Mab (Jiangsu, China) for providing the spike proteins of Alpha, Delta, and Lambda, and the hACE2 protein.
Disclosure
Liang Ye and Dandan Wu report a pending patent on the combination of Nb-H6 mentioned in the paper. Chinese patent application No. 202210681887.X. The authors declare that they have no other competing interests in this work.
References
1. Beaudoin-Bussieres G, Laumaea A, Anand SP, et al. Decline of Humoral Responses against SARS-CoV-2 Spike in Convalescent Individuals. mBio. 2020;11(5). doi:10.1128/mBio.02590-20.
2. Kustin T, Harel N, Finkel U, et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat Med. 2021;27(8):1379–1384. doi:10.1038/s41591-021-01413-7
3. Becker M, Dulovic A, Junker D, et al. Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat Commun. 2021;12(1):3109. doi:10.1038/s41467-021-23473-6
4. Peng Q, Zhou R, Wang Y, et al. Waning immune responses against SARS-CoV-2 variants of concern among vaccinees in Hong Kong. EBioMedicine. 2022;77:103904. doi:10.1016/j.ebiom.2022.103904
5. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi:10.1038/s41586-020-2180-5
6. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292 e6. doi:10.1016/j.cell.2020.02.058
7. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280 e8. doi:10.1016/j.cell.2020.02.052
8. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183(6):1735. doi:10.1016/j.cell.2020.11.032
9. Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: implications for the Design of Spike-Based Vaccine Immunogens. Front Immunol. 2020;11:576622. doi:10.3389/fimmu.2020.576622
10. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi:10.1126/science.abc6952
11. Wu Y, Li C, Xia S, et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27(6):891–898 e5. doi:10.1016/j.chom.2020.04.023
12. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi:10.1038/s41586-020-2381-y
13. Wagner TR, Ostertag E, Kaiser PD, et al. NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies. EMBO Rep. 2021;22(5):e52325. doi:10.15252/embr.202052325
14. Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi:10.1146/annurev-biochem-063011-092449
15. Konwarh R. Nanobodies: prospects of Expanding the Gamut of Neutralizing Antibodies Against the Novel Coronavirus, SARS-CoV-2. Front Immunol. 2020;11:1531. doi:10.3389/fimmu.2020.01531
16. Haga K, Takai-Todaka R, Matsumura Y, et al. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17(10):e1009542. doi:10.1371/journal.ppat.1009542
17. Laursen NS, Friesen RHE, Zhu X, et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362(6414):598–602. doi:10.1126/science.aaq0620
18. Jiang S, Hillyer C, Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020;41(5):355–359. doi:10.1016/j.it.2020.03.007
19. Li D, Sempowski GD, Saunders KO, Acharya P, Haynes BF. SARS-CoV-2 Neutralizing Antibodies for COVID-19 Prevention and Treatment. Annu Rev Med. 2022;73:1–16. doi:10.1146/annurev-med-042420-113838
20. Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2(11):3001–3008. doi:10.1038/nprot.2007.448
21. Schoof M, Faust B, Saunders RA, et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370(6523):1473–1479. doi:10.1126/science.abe3255
22. Huo JD, Le Bas A, Ruza RR, et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27(9):846–+. doi:10.1038/s41594-020-0469-6
23. Li Q, Zhang F, Lu Y, et al. Highly potent multivalent VHH antibodies against Chikungunya isolated from an alpaca naive phage display library. J Nanobiotechnology. 2022;20(1):231. doi:10.1186/s12951-022-01417-6
24. Cabanas-Danes J, Rodrigues ED, Landman E, et al. A supramolecular host-guest carrier system for growth factors employing V(H)H fragments. J Am Chem Soc. 2014;136(36):12675–12681. doi:10.1021/ja505695w
25. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi:10.1038/s41586-021-03819-2
26. Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic fab libraries with a single framework scaffold. J Mol Biol. 2004;340(5):1073–1093. doi:10.1016/j.jmb.2004.05.051
27. Barnes CO, Jette CA, Abernathy ME, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi:10.1038/s41586-020-2852-1
28. Huo J, Le Bas A, Ruza RR, et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27(9):846–854. doi:10.1038/s41594-020-0469-6
29. Tang Q, Owens RJ, Naismith JH. Structural Biology of Nanobodies against the Spike Protein of SARS-CoV-2. Viruses. 2021;13(11):67.
30. Li D, Edwards RJ, Manne K, et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184(16):4203–4219 e32. doi:10.1016/j.cell.2021.06.021
31. Shimizu J, Sasaki T, Koketsu R, et al. Reevaluation of antibody-dependent enhancement of infection in anti-SARS-CoV-2 therapeutic antibodies and mRNA-vaccine antisera using FcR- and ACE2-positive cells. Sci Rep. 2022;12(1):15612. doi:10.1038/s41598-022-19993-w
32. Wang S, Wang J, Yu X, et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcgammaRIIB and virus-antibody complex with bivalent interaction. Commun Biol. 2022;5(1):262. doi:10.1038/s42003-022-03207-0
33. Okuya K, Hattori T, Saito T, et al. Multiple Routes of Antibody-Dependent Enhancement of SARS-CoV-2 Infection. Microbiol Spectr. 2022;10(2):e0155321. doi:10.1128/spectrum.01553-21
34. Maemura T, Kuroda M, Armbrust T, Yamayoshi S, Halfmann PJ, Kawaoka Y. Antibody-Dependent Enhancement of SARS-CoV-2 Infection Is Mediated by the IgG Receptors FcgammaRIIA and FcgammaRIIIA but Does Not Contribute to Aberrant Cytokine Production by Macrophages. mBio. 2021;12(5):e0198721. doi:10.1128/mBio.01987-21
35. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20(6):339–341. doi:10.1038/s41577-020-0321-6
36. Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol. 2020;5(10):1185–1191. doi:10.1038/s41564-020-00789-5
37. Liu Y, Soh WT, Kishikawa JI, et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184(13):3452–3466 e18. doi:10.1016/j.cell.2021.05.032
38. Shimizu J, Sasaki T, Yamanaka A, et al. The potential of COVID-19 patients’ sera to cause antibody-dependent enhancement of infection and IL-6 production. Sci Rep. 2021;11(1):23713. doi:10.1038/s41598-021-03273-0
39. Arvin AM, Fink K, Schmid MA, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–363. doi:10.1038/s41586-020-2538-8
40. Zang J, Gu C, Zhou B, et al. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020;6:61. doi:10.1038/s41421-020-00199-1
41. Li JF, He L, Deng YQ, et al. Generation and Characterization of a Nanobody Against SARS-CoV. Virol Sin. 2021;36(6):1484–1491. doi:10.1007/s12250-021-00436-1
42. Hanke L, Vidakovics Perez L, Sheward DJ, et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11(1):4420. doi:10.1038/s41467-020-18174-5
43. Chi X, Zhang X, Pan S, et al. An ultrapotent RBD-targeted biparatopic nanobody neutralizes broad SARS-CoV-2 variants. Signal Transduct Target Ther. 2022;7(1):44. doi:10.1038/s41392-022-00912-4
44. Weinstein JB, Bates TA, Leier HC, McBride SK, Barklis E, Tafesse FG. A potent alpaca-derived nanobody that neutralizes SARS-CoV-2 variants. iScience. 2022;25(3):103960. doi:10.1016/j.isci.2022.103960
45. Lu Q, Zhang Z, Li H, et al. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J Nanobiotechnology. 2021;19(1):33. doi:10.1186/s12951-021-00768-w
46. Xiang Y, Nambulli S, Xiao Z, et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–1484. doi:10.1126/science.abe4747
47. Wagner TR, Schnepf D, Beer J, et al. Biparatopic nanobodies protect mice from lethal challenge with SARS-CoV-2 variants of concern. EMBO Rep. 2022;23(2):e53865. doi:10.15252/embr.202153865
48. Yang Q, Jian X, Syed AAS, et al. Structural Comparison and Drug Screening of Spike Proteins of Ten SARS-CoV-2 Variants. Research. 2022;2022:9781758. doi:10.34133/2022/9781758
49. Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi:10.1038/s41579-021-00573-0
50. Sungnak W, Huang N, Becavin C, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. doi:10.1038/s41591-020-0868-6
51. Zhou R, Wang P, Wong YC, et al. Nasal prevention of SARS-CoV-2 infection by intranasal influenza-based boost vaccination in mouse models. EBioMedicine. 2022;75:103762. doi:10.1016/j.ebiom.2021.103762
52. Froberg J, Gillard J, Philipsen R, et al. SARS-CoV-2 mucosal antibody development and persistence and their relation to viral load and COVID-19 symptoms. Nat Commun. 2021;12(1):5621. doi:10.1038/s41467-021-25949-x
53. Fan W, Sun S, Zhang N, et al. Nasal delivery of thermostable and broadly neutralizing antibodies protects mice against SARS-CoV-2 infection. Signal Transduct Target Ther. 2022;7(1):55. doi:10.1038/s41392-022-00911-5
54. Halwe S, Kupke A, Vanshylla K, et al. Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses. 2021;13(8):87.
55. Chen RE, Winkler ES, Case JB, et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596(7870):103–108. doi:10.1038/s41586-021-03720-y
56. Nambulli S, Xiang Y, Tilston-Lunel NL, et al. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci Adv. 2021;7(22):45.
57. Li C, Zhan W, Yang Z, et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185(8):1389–1401 e18. doi:10.1016/j.cell.2022.03.009
58. Esparza TJ, Chen Y, Martin NP, et al. Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19. MAbs. 2022;14(1):2047144. doi:10.1080/19420862.2022.2047144
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.