Back to Journals » OncoTargets and Therapy » Volume 11
A 5-serum miRNA panel for the early detection of colorectal cancer
Authors Guo S, Zhang J, Wang B, Zhang B, Wang X, Huang L, Liu H, Jia B
Received 9 October 2017
Accepted for publication 20 March 2018
Published 8 May 2018 Volume 2018:11 Pages 2603—2614
DOI https://doi.org/10.2147/OTT.S153535
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Ingrid Espinoza
Shaohua Guo,1,2 Jiajin Zhang,2 Baishi Wang,2 Bingdong Zhang,1 Xuning Wang,1 Liang Huang,1 Hongyi Liu,2 Baoqing Jia1,2
1Chinese PLA Medical School, Beijing, China; 2General Surgery Department II, Chinese PLA General Hospital, Beijing, China
Objective: The study aimed to screen microRNAs (miRNAs) that can be used for the early detection of colorectal cancer (CRC) based on differential expression of miRNA in serum.
Materials and methods: A three-stage study was designed with a total of 217 CRCs, 168 colorectal adenomas (CRAs), and 190 healthy controls (HCs). A quantitative reverse transcription polymerase chain reaction was performed in three stages. We screened 528 miRNA expression profiles in the sera of 40 patients (CRC n=20, CRA n=10, and HC n=10) for candidate miRNAs, then 210 serum samples (CRC n=90, CRA n=60, and HC n=60) were used for screening of candidate miRNAs. Three hundred and twenty-five independent individual samples (CRC n=107, CRA n=98, and HC n=120) were used to validate the most differentially-expressed miRNAs in the screening stage, and binary logistic regression was used in the validation stage. A receiver operating characteristic curve was drawn to evaluate the diagnostic accuracy.
Results: A 5-serum miRNA panel (miRNA-1246, miRNA-202-3p, miRNA-21-3p, miRNA-1229-3p, and miRNA-532-3p) effectively distinguished CRCs from HCs with 91.6% sensitivity and 91.7% specificity. The area under the curve (AUC) was 0.960 (95% confidence interval [CI]: 0.937–0.983). In addition, the panel also accurately distinguished CRCs from CRAs with 94.4% sensitivity and 84.7% specificity. The AUC was 0.951 (95% CI: 0.922–0.980).
Conclusion: Our 5-serum miRNA panel accurately distinguished CRCs from CRAs and HCs with high sensitivity and specificity. The 5-serum miRNA panel may be a promising prospect for application as a nonintrusive and inexpensive method for the early detection of CRC.
Keywords: colorectal cancer, colorectal adenoma, miRNA
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors. The incidence of CRC among malignant tumors in males and females ranks third and second, respectively. In fact, 1.3 million people are diagnosed with CRC and there are nearly 700,000 deaths annually worldwide; CRC ranks fourth among cancer-related deaths.1 In China, the number of new cases of CRC in 2015 exceeded 3,763,000 and the number of CRC-specific deaths was approximately 1,910,000.2 In recent years, with the improvement in living standards and Westernization of lifestyles, the incidence and mortality of CRC have shown a rising trend year after year. Indeed, most patients with CRC are in an advanced stage at the time of diagnosis and have lost curability. The current study showed that the mortality rate of CRC was highly correlated with the stage of disease. The 5-year survival rate for stage I CRC approaches 90%, but is only 12% for stage IV.3 It is evident that early detection of CRC and appropriate treatment are important. Currently, the common methods for early detection of CRC include the plasma carcinoembryonic antigen (CEA) level, fecal occult blood testing, and fiberoptic colonoscopy; however, the plasma CEA level and fecal occult blood testing are not highly sensitive, and therefore their value is limited.4–6 Colonoscopy is considered the gold standard for detection of CRC; however, colonoscopy is still limited as an effective approach for early detection due to poor patient compliance, resulting from discomfort during bowel preparation and the examination, as well as the potential complications.7 Therefore, a new method which is less invasive, highly accurate, and of clinical value for the early detection of CRC is warranted.
MicroRNAs (miRNAs) are a class of noncoding single-stranded RNA molecules (approximately 21–23 nucleotides in length) coded by endogenous genes and specifically bound to the 3' untranslated region of target mRNAs to inhibit translation and promote degradation.8 miRNAs participate in a variety of vital biological processes, including cell differentiation, proliferation, and apoptosis. The mechanism of its action may be related to miRNA-related single nucleotide polymorphisms.9–12 It has been reported that abnormally-expressed miRNA, as an oncogene or tumor suppressor, plays an important role in cancer initiation and progression, including lung, liver, pancreatic, bladder, gastric, ovarian, and colorectal cancers.13–17 There are a variety of miRNAs in human blood that are usually derived from lysis of apoptotic or necrotic cells and secretion of specific cells.18 miRNAs remain stable by binding to Argonaute 2 protein19 or through the autospecific hairpin structure,20 which prevents degradation by ribonuclease in the blood and facilitates quantitative detection. In 2008, Lawrie et al21 first reported the quantitative detection of miRNA in serum samples, proving the feasibility for the approach. An increasing number of studies on various diseases, especially on tumors, have focused on circulation miRNA.22,23 However, in our study, more samples were included and the method of quantitative reverse transcription polymerase chain reaction (qRT-PCR) cost less. The objective of the current study was to identify differentially-expressed miRNAs and to determine the feasibility of their use in the early detection of CRC.
Materials and methods
Study subjects
The study was approved by the Ethics Committee of the Chinese PLA General Hospital, and all patients signed written informed consent forms. The inclusion criteria were as follows: (1) colonoscopy; (2) no inflammatory bowel diseases, hereditary adenomatous polyposis, family history of tumors, and other tumors in the preceding or corresponding periods; (3) colorectal adenomas (CRAs) and CRCs should have pathologic confirmation; and (4) no preoperative chemotherapy for patients with CRC. The patients with CRC were staged according to the TNM staging system of the American Joint Committee on Cancer.49 A total of 575 patients were enrolled from the Department of Gastrointestinal Surgical Oncology, Department of Gastroenterology, and Medical Examination Center of the Chinese PLA General Hospital between April 2013 and December 2014. Their general statistical information is presented in Table 1. The collection of all blood samples followed a uniform protocol, ie, a 5-mL vacuum blood collection tube (VACUETTE®; Greiner Bio-One, Frickenhausen, Germany) was used to collect blood, and the sample was allowed to stand for 2 h at room temperature to fully coagulate, and then centrifuged at 1,500 rpm (2,390×g) at 4°C for 7 min. The centrifugation was completed within 4 h after blood collection. The serum samples collected after centrifugation were stored in a −80°C refrigerator.
  | Table 1 Clinical characteristics of patients |
Study design
Our study was divided into three stages (primary screening, screening, and validation). The study design is shown in Figure 1.
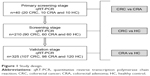  | Figure 1 Study design. |
Primary screening and screening stages
During the primary screening stage, we screened 528 miRNA expression profiles in 40 serum samples, and CRCs, CRAs, and healthy controls (HCs) were compared in pairs. One hundred and five miRNAs were differentially expressed (Tables S1–S3). The primary screening criteria for differentially-expressed miRNAs were as follows: (1) mean cycle threshold (Ct) <32 and SD within groups <2; (2) 5 Cts less than the negative control (no template control); and (3) the Ct value of the same sample was standardized on the same plate with an external reference (cel-miR-67) to obtain the ΔCt value for comparison between groups and to select the most differentially-expressed miRNAs.
The serum sample size was expanded (n=210) and candidate miRNAs (105 miRNAs in the primary screening stage) satisfying the above three criteria for primary screening were rescreened. The screening criteria were as follows: (1) the results of primary screening and screening were compared, and miRNAs with consistent expression patterns or mean Ct differences between groups >1 were selected; (2) mean Ct <30; and (3) the Ct value of the same sample was standardized on the same plate with an external reference (cel-miR-67) to obtain the ΔCt value for comparison between groups and to select the most differentially-expressed miRNAs. Validation was conducted on miRNAs which satisfied all three screening criteria.
Validation stage
During the screening stage, we identified 10 most significant miRNAs. There were 5 miRNAs in group of HC vs CRA, 9 miRNAs in groups of CRA vs HC, and 9 miRNAs in group of CRC vs HC. But some miRNAs are repetitive (Table S4, Tables 2 and 3). Three hundred and twenty-five serum samples (CRC n=107, CRA n=98, and HC n=120) were used to validate the difference and binary logistic regression was based on data from the validation stage. We validated the 5 miRNAs with the most significant difference (Figure S1). A receiver operating characteristic curve was drawn and a relevant mathematical model was established for detection of CRC.
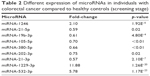  | Table 2 Different expression of microRNAs in individuals with colorectal cancer compared to healthy controls (screening stage) |
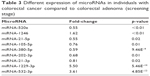  | Table 3 Different expression of microRNAs in individuals with colorectal cancer compared to colorectal adenoma (screening stage) |
Extraction of serum total RNA
During extraction of total RNA, 1 μL of cel-miR-67 (20 nM/L; Quantobio, Beijing, China) was added to each sample as an external reference. A total of 0.25 mL of serum was used in the extraction of total RNA following the procedures in the miRNA extraction kit (Biochain, Beijing, China). A Nanodrop2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the quality of total RNA and the OD260-to-OD280 ratio was within 1.8–2.1.
Quantitative reverse transcription polymerase chain reaction
During the screening and validation stages, we used a three-step qRT-PCR. PolyA polymerase (NEB, Beijing, China) was used to add an A tail for the miRNA primer and mature miRNA. The reaction system was in a volume of 20 μL at 37°C for 1 h. Then, cRNA was synthetized by reverse transcription. One microliter of universal reverse transcription primer (Quantobio) of the tested 528 miRNAs was added to each reaction tube and incubated at 70°C for 5 min, 4°C for 2–10 min, 50°C for 50 min, and 70°C for 15 min. cDNA was obtained. qPCR was performed on an ABI 7900 High Throughput Rapid Real-time Fluorescence Quantitative PCR Amplifier (Applied Biosystems; Thermo Fisher Scientific) at 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 45 s. The melting analysis was added to evaluate the specificity of the PCR products: 95°C for 15 s, 60°C for 15 s, and then 95°C for 15 s. Quantification of the PCR product was evaluated by the level of fluorescence emitted by SYBR Green (SYBR® Premix Ex Taq™ II; TaKaRa, Beijing, China).
Statistical analysis
The ΔCt value was used to represent the relative level of expression of a single miRNA. A two-tailed t-test was used between two independent samples to identify significantly-expressed miRNA. Binary logistic regression was used during the validation stage and a receiver operating characteristic curve was drawn to calculate the area under the curve (AUC) and assess the diagnostic value of miRNA for CRC. Based on relevant parameters in binary logistic regression, we established a mathematical diagnosis equation for detection of CRC. SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA) was used to perform all statistical analyses. Differences were considered significant at p<0.05.
Results
Comparison between CRCs and HCs
As compared with HCs, there was a significant difference in 9 miRNAs in the sera of patients with CRC during the screening stage (Table 2). After validation, a panel of 5 miRNAs (miRNA-1246, miRNA-202-3p, miRNA-21-3p, miRNA-1229-3p, and miRNA-532-3p) effectively distinguished CRCs (n=107) from HCs (n=120), with 91.6% sensitivity and 91.7% specificity. The AUC was 0.960 (95% confidence interval [CI]: 0.937–0.983) (Figure 2). Based on relevant parameters in binary logistic regression, a diagnosis discriminant model was established according to the following equation:
|
The cutoff value was 0.502. When Y1<0.502, the detection result was negative, and determined to be a HC. When Y1≥0.502, the detection result was positive, and determined to be a CRC.
Comparison between CRCs and CRAs
As compared with the CRA group, there was a significant difference in 9 miRNAs in the sera of patients with CRCs during the screening stage (Table 3). After validation, a panel of 5 miRNAs (miRNA-1246, miRNA-202-3p, miRNA-21-3p, miRNA-1229-3p, and miRNA-532-3p) effectively distinguished CRCs (n=107) from CRAs (n=98), with 94.4% sensitivity and 84.7% specificity. The AUC was 0.951 (95% CI: 0.922–0.980) (Figure 3). Based on relevant parameters in binary logistic regression, a diagnosis discriminant model was established according to the following equation:
|
The cutoff value was 0.348. When Y2<0.348, the detection result was negative, and determined to be a CRA. When Y2≥0.348, the detection result was positive, and determined to be a CRC.
Discussion
The incidence of CRC is increasing year after year. A former study revealed that good prognosis can be achieved for the majority of CRC patients if there is early detection and appropriate treatment.24 Therefore, early detection becomes key for disease prophylaxis. In recent years, circulating miRNAs have attracted increased attention from investigators, which have become markers for early tumor detection. Chen et al25 reported that serum miRNA expression profiling of patients with CRC differs from the healthy population, and believed that part of the abnormally-expressed miRNA could be used for detection of CRC. Ng et al26 reported that in patients with CRC, the level of miRNA-92a expression is significantly increased not only in cancer tissues, but in plasma as well. After resection of cancerous tissue during surgery, however, plasma miRNA-92a is restored to the normal level and it was confirmed that high miRNA-92a expression is unrelated to inflammatory bowel disease and gastric cancer. Huang et al27 monitored the levels of miRNA-92a expression in 20 CRC patients before and after surgery. The results indicated an obvious decrease after surgery, which was consistent with the findings of Ng et al,26 further confirming the value of miRNA92a in detection of CRC. Toiyama et al28 tested abnormally-expressed miRNA in serum samples of patients with CRCs, CRAs, and HCs, and found that the level of miR-21 expression in sera of patients with CRAs and CRCs before surgery was increased and significantly decreased after surgery. They concluded that miR-21 is a potential marker for the early detection of CRC. Kanaan et al29 reported that the sensitivity and specificity of distinguishing CRC from the control group based on increased miR-21 expression in plasma were up to 90%; however, miR-92a and miR-21 had poor organ specificity: miRNA-92a was increased in patients with acute leukemia and cervical cancer;30,31 and miR-21 was also highly expressed in patients with lung, breast, esophageal, and gastric cancers.32 Therefore, it may be difficult to assess results when a single miRNA is used as a diagnostic marker. More and more investigators have proposed a combination of several miRNAs to improve diagnostic efficiency. Some studies have also confirmed the feasibility of a panel consisting of multiple circulating miRNAs in the early detection of CRC. Luo et al33 showed that 9 differentially-expressed miRNAs (miR-18a, miR-20a, miR-21, miR-29a, miR-92a, miR-106b, miR-133a, miR-143, and miR-145) are present in the plasma of patients with CRC. The 9-miRNA panel accurately distinguished patients with CRC from patients with non-CRC, with an AUC of 0.745. Wang et al34 reported that a panel consisting of miR-409-3p, miR-7, and miR-93 accurately diagnosed CRC with a sensitivity of 82%, specificity of 89%, and AUC of 0.897. Zheng et al35 believed that a 4-serum miRNA panel (miR-19a-3p, miR-223-3p, miR-92a-3p, and miR-422a) diagnosed early CRC with an AUC up to 0.951. Moreover, the panel accurately identified CRCs, CRAs, and HCs with AUCs of 0.886 and 0.765, respectively. Zhu et al36 showed that the serum levels of miR-19a-3p, miR-21-5p, and miR-425-5p expression in patients with CRC were higher than normal controls. The panel could be used in detection of CRC with an AUC of 0.830. Nevertheless, there are currently no relevant diagnostic products widely available for clinical use, probably because of the variety of races and samples, and different detection approaches. Therefore, we suggest that additional research data are warranted to provide theoretical support for clinical use.
A total of 575 serum samples were collected in this study, including 217 CRCs, 168 CRAs, and 190 HCs. qRT-PCR was used in all three stages. Through screening and validation, a 5-serum miRNA panel (miRNA-1246, miRNA-202-3p, miRNA-21-3p, miRNA-1229-3p, and miRNA-532-3p) was confirmed to differentiate CRCs from HCs, and CRCs from CRAs. The AUCs were 0.960 and 0.951, respectively, which were superior to previous studies.33–38 As compared with the CRC group, there was a significant difference in the serum level of expression of 5 miRNAs in the HC and CRA groups. All new diagnostic tests have limitations, especially pertaining to the sample size of the study. A total of 575 serum samples were included in our study, which is larger than other studies, thus ensuring the reliability of the test results. In addition, we adopted a three-step qRT-PCR to avoid the false-negative results in the two-step method caused by stem-loop primers, and adopted a mash-up of long and short primers to ensure high efficiency and accuracy of the amplification. Finally, we also established a mathematical model for CRC detection, which provides great convenience for future clinical application.
Proper standardization is crucial for accurate quantitation of miRNA expression by qRT-PCR. Kanaan et al39 reported that a panel of 3 plasma miRNAs (miR-431, miR-15b, and miR-139-3p) distinguished stage IV CRC from controls and successfully used U6 as an internal control to standardize miRNA expression. Rice et al40 recommended using RNU6 and miR-520d-5p as reliable internal controls in a study of plasma miRNA; however, some reports hold the view that U6 is not expressed constantly and easily degraded by RNase in the circulation; the stability is poor.25,41,42 To date, no uniform standard for internal control has been established. Therefore, it has been proposed to use an external reference for standardization of the level of circulating miRNA expression.43 Wang et al44 concluded that the miRNA (cel-miR-39) of Caenorhabditis elegans is nonhomologous with humans and has good repeatability and high stability. Therefore, cel-miR-39 can be used for quantification of circulating miRNA by qRT-PCR. Zhu et al36 reported that the serum levels of miR-19a-3p, miR-21-5p, and miR-425-5p expression in patients with CRC are significantly higher than in the control group. They also used cel-miR-39 as an external reference for standardized calculation of the relative expression of miRNAs. Cel-miR-67 is a miRNA of C. elegans. It has no homology with human miRNA and meets the requirement of external reference.45 We successfully used cel-miR-67 (primer sequence, CAACCTCCTAGAAAGAGTA) as an external reference and obtained stable and repeatable results.
Wang et al46 reported that upregulation of miRNA-1246 in CRC tissues may cause disease progression. The cause can be miRNA-1246 binding to CCNG2 in CRC cells, which inhibits the expression of cyclin 2 (CycG2). Lai and Friedman47 detected that miRNA-1246 is significantly increased in the sera of patients with CRC and suggested that miRNA-1246 could be used as a potential biomarker for early detection of CRC. Wang et al48 showed low expression of miRNA-202-3p in cancer tissues in patients with CRC and demonstrated overexpression of miRNA-202-3p inhibited growth of CRC cells in vitro. However, we have not uncovered any reports that the three miRNAs (miR-21-3p, miR-532-3p, and miR-1229-3p) serve as markers for the early detection of CRC, as reported for the first time in the current study.
Conclusion
Circulating miRNAs can be considered to be important biomarkers for detection of CRC. We found 5 differentially-expressed miRNAs in serum, and as a non-invasive screening approach for CRC, with high sensitivity and specificity, the panel may be used for accurate early detection of CRC, which is a promising prospect for clinical application.
Acknowledgments
We thank Ms Xinxin Liu and Mr Gaoshang Song for their support. This work was supported by the 863 project of China (No 2014AA020802).
Disclosure
The authors report no conflicts of interest in this work.
References
McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
El-Shami K, Oeffinger KC, Erb NL, et al. American Cancer Society Colorectal Cancer Survivorship Care Guidelines. CA Cancer J Clin. 2015;65(6):428–455. | ||
Fakih MG, Padmanabhan A. CEA monitoring in colorectal cancer. What you should know. Oncology (Williston Park). 2006;20(6):579–587; discussion 588, 594, 596 passim. | ||
Young GP, Symonds EL, Allison JE, et al. Advances in Fecal Occult Blood Tests: the FIT revolution. Dig Dis Sci. 2015;60(3):609–622. | ||
Lieberman D. Screening, surveillance and prevention of colorectal cancer. Gastrointest Endosc Clin N Am. 2008;18(3):595–605. | ||
Nicholson FB, Korman MG. Acceptance of flexible sigmoidoscopy and colonoscopy for screening and surveillance in colorectal cancer prevention. J Med Screen. 2005;12(2):89–95. | ||
Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. | ||
Gopalakrishnan C, Kamaraj B, Purohit R. Mutations in microRNA binding sites of CEP genes involved in cancer. Cell Biochem Biophys. 2014;70(3):1933–1942. | ||
Bhaumik P, Gopalakrishnan C, Kamaraj B, Purohit R. Single nucleotide polymorphisms in microRNA binding sites:implications in colorectal cancer. Scientific WorldJournal. 2014;2014:547154. | ||
Kamaraj B, Gopalakrishnan C, Purohit R. In silico analysis of miRNA-mediated gene regulation in OCA and OA genes. Cell Biochem Biophys. 2014;70(3):1923–1932. | ||
Kumar A, Purohit R. Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Comput Biol. 2014;10(4):e1003318. | ||
Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435(7043):745–746. | ||
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. | ||
Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. | ||
Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. | ||
Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. | ||
Lawrie AS, Cardigan RA, Williamson LM, Machin SJ, Mackie IJ. The dynamics of clot formation in fresh-frozen plasma. Vox Sang. 2008;94(4):306–314. | ||
Chevillet JR, Lee I, Briggs HA, He Y, Wang K. Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules. 2014;19(5):6080–6105. | ||
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. | ||
Akkoca AN, Yanik S, Ozdemir ZT, et al. TNM and Modified Dukes staging along with the demographic characteristics of patients with colorectal carcinoma. Int J Clin Exp Med. 2014;7(9):2828–2835. | ||
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. | ||
Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. | ||
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–126. | ||
Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. | ||
Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–551. | ||
Ohyashiki JH, Umezu T, Kobayashi C, et al. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: in vivo assessment of cell to plasma ratio of miR-92a. BMC Res Notes. 2010;3:347. | ||
Kong Q, Tang Z, Xiang F, et al. Diagnostic value of serum hsa-mir-92a in patients with cervical cancer. Clin Lab. 2017;63(2):335–340. | ||
Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138(10):1659–1666. | ||
Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8(5):e62880. | ||
Wang S, Xiang J, Li Z, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136(1):152–161. | ||
Zheng G, Du L, Yang X, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111(10):1985–1992. | ||
Zhu M, Huang Z, Zhu D, et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget. 2017;8(10):17081–17091. | ||
Carter JV, Roberts HL, Pan J, et al. A highly predictive model for diagnosis of colorectal neoplasms using plasma microRNA: improving specificity and sensitivity. Ann Surg. 2016;264(4):575–584. | ||
Chen WY, Zhao XJ, Yu ZF, et al. The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int J Clin Exp Pathol. 2015;8(6):7092–7101. | ||
Kanaan Z, Roberts H, Eichenberger MR, et al. A plasma microRNA panel for detection of colorectal adenomas:a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258(3):400–408. | ||
Rice J, Roberts H, Rai SN, Galandiuk S. Housekeeping genes for studies of plasma microRNA: a need for more precise standardization. Surgery. 2015;158(5):1345–1351. | ||
Chen X, Liang H, Guan D, et al. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLOS One. 2013;8(11):e79652. | ||
Wulfken LM, Moritz R, Ohlmann C, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6(9):e25787. | ||
Liu ZL, Slininger PJ. Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods. 2007;68(3):486–496. | ||
Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7(9):e44398. | ||
Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. | ||
Wang S, Zeng Y, Zhou JM, et al. MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol Med Rep. 2016;13(1):273–280. | ||
Lai X, Friedman A. Exosomal microRNA concentrations in colorectal cancer: a mathematical model. J Theor Biol. 2017;415:70–83. | ||
Wang Q, Huang Z, Guo W, et al. MicroRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Clin Cancer Res. 2014;20(5):1146–1157. | ||
Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. |
Supplementary materials
  | Table S1 Different expression of microRNAs in individuals with colorectal cancer compared to colorectal adenoma (primary screening stage) |
  | Table S2 Different expression of microRNAs in individuals with colorectal adenoma compared to healthy controls (primary screening stage) |
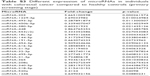  | Table S3 Different expression of microRNAs in individuals with colorectal cancer compared to healthy controls (primary screening stage) |
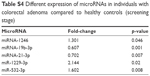  | Table S4 Different expression of microRNAs in individuals with colorectal adenoma compared to healthy controls (screening stage) |
  | Figure S1 The 5 microRNAs have different expression in the 3 groups. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




