Back to Journals » OncoTargets and Therapy » Volume 9
WISP3 is highly expressed in a subset of colorectal carcinomas with a better prognosis
Authors Lu Y, Wang X, Sun X, Feng W, Guo H, Tang C, Anmeideng A, Bao Y
Received 24 September 2015
Accepted for publication 15 November 2015
Published 12 January 2016 Volume 2016:9 Pages 287—293
DOI https://doi.org/10.2147/OTT.S97025
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Yongliang Lu,1,* Xiang Wang,2,* Xinrong Sun,2 Wenming Feng,2 Huihui Guo,2 Chengwu Tang,2 Anmei Deng,3 Ying Bao2
1Department of Medicine, Huzhou Teachers College, 2Department of Gastrointestinal Surgery, First Affiliated Hospital, Huzhou Teachers College, The First People’s Hospital of Huzhou, Huzhou, 3Department of Laboratory Diagnostic, Changhai Hospital, Second Military Medical University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Outlier genes with marked overexpression in subsets of cancers like ERBB2 have potential for the identification of gene classifiers and therapeutic targets for the appropriate subpopulation. In this study, using the cancer outlier profile analysis strategy, we identified WNT1-inducible-signaling pathway protein 3 (WISP3) as an outlier gene that is highly expressed in a subset of colorectal cancers (CRCs) from The Cancer Genome Atlas dataset. A meta-cancer outlier profile analysis and immunohistochemistry experiment to validate the outlier expression model of WISP3 in CRC was then performed. Our immunohistochemical results indicated that WISP3 was more frequently seen in the small tumors, and there was a significant association between its overexpression with a good prognosis. Furthermore, in the multivariable model, WISP3 outlier expression retained significance for overall survival. In summary, in this study, we identified an outlier gene WISP3 overexpressed in a subset of CRC having less aggressive characteristics and a better prognosis. We suggest WISP3 may provide more accurate and precise information regarding CRC population classification.
Keywords: subtype, WISP3, outlier, prognosis, microarray
Introduction
Identification of gene aberrations is of important biological and clinical significance for cancer research. The most common strategy was to compare gene expression profiles between cancerous and normal samples to identify consistently aberrantly expressed genes in tumor samples. However, the limitation with such an approach is that it ignores the tumor heterogeneity, and fails to find outlier genes, which is overexpressed only in a subgroup of tumors, but not at the whole level.1 Meanwhile, many clinically validated tumor biomarkers belong to outlier oncogenes, for example, ERBB2 is overexpressed only in 15%–20% of breast tumors while it shows a modest elevation within breast cancers compared with normal breast tissue.2
Colorectal cancer (CRC) is a typically heterogeneous disease, and demonstrated a relatively high incidence and poor clinical outcome.3 Although several biomarkers, such as microsatellite instability, carcinoembryonic antigen, and k-ras mutation testing, have been clinically used for the management of CRC, their benefits are only limited to a small number of patients. Therefore, there is a need to develop novel biomarkers in clinical practice for early diagnosis, disease monitoring, and predicting prognosis and therapy response.
Identification of outlier genes overexpressed in a subset of samples has been proven to be successful in finding critical gene classifiers and prognostic factors.4 Until now, only a few studies have examined the heterogeneity that exists between individual CRC cancers. For example, using high throughput RNA sequencing and outlier expression analysis, a recent study identified a novel transcript of VNN1-AB gene expressed in a subset of CRC samples.5
In this study, we performed a cancer outlier profile analysis (COPA) to identify novel outlier genes specific for a subset of CRC tumors from The Cancer Genome Atlas (TCGA) gene expression data. Our analysis nominates WNT1-inducible-signaling pathway protein 3 (WISP3) as an outlier gene that is highly expressed in a subset of CRC tumors across independent cohorts. We also experimentally confirmed that WISP3 expression in CRC was associated with a better prognosis.
Materials and methods
Gene outlier expression analyses from TCGA CRC mRNA dataset
COPA was performed on TCGA CRC mRNA expression dataset from the Oncomine database as described previously.6 COPA function has been implemented in the Oncomine database (https://www.oncomine.org). TCGA CRC mRNA expression dataset in the Oncomine database included 215 colorectal adenocarcinoma and 22 paired normal colorectal tissue samples. TCGA mRNA expression data were produced on Agilent 244K Custom Gene Expression microarray platform (Agilent Technologies, Santa Clara, CA, USA) and Illumina RNA-Seq platform (Illumina, Inc., San Diego, CA, USA). Samples from the TCGA CRC mRNA dataset are scored based on rescaled median absolute deviation, and COPA scores are calculated at 90th and 75th percentiles. Then genes are rank-ordered based on 90th and 75th percentile scores.
Meta-COPA analysis of WISP3 in CRC datasets
We selected the top outlier gene WISP3 identified from TCGA CRC mRNA dataset for further meta-COPA analysis in other three independent CRC microarray cohorts (Vilar Colorectal 2, Vilar Colorectal, and Smith Colorectal) as described previously.7,8 All these three validation datasets were performed on Affymetrix Human Genome Array platforms. Vilar Colorectal 2, Vilar Colorectal, and Smith Colorectal datasets included 176, 155, and 177 CRC samples; no normal control tissue was included in these three cohorts.
CRC patients and specimens
A total of 185 CRC patients who underwent surgical resection were included in this study and provided written informed consent. All the patients have adequate volume of formalin-fixed paraffin-embedded tumor specimens. The patients received treatment according to the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines of Chinese version. The follow-up, which is defined as the time between surgical resection and death, ranged from 1 to 97 months. The procedures of this study were approved by the research ethics committee of The First People’s Hospital of Huzhou.
Immunohistochemistry
Immunohistochemistry was performed using the streptavidin–biotin–peroxidase system. Briefly, 4 μm of sections were deparaffinized, and endogenous peroxidases were quenched with 3% H2O2. After microwave-citrate antigen retrieval in 10 mM citrate buffer (pH 6.0) for 1 hour, sections were incubated with rabbit antibodies antihuman WISP3 (1:100; Abcam, Cambridge, MA, USA) overnight at 4°C. Staining was subsequently localized by using diaminobenzidine tetrahydrochloride as a chromogen, and was then counterstained with hematoxylin. For negative controls, WISP3 antibody was replaced by nonspecific rabbit immunoglobulin G.
The results of WISP3 immunostaining was semiquantified using H score system by multiplying the percentage of staining tumor cells (1, <10%; 2, 10%–30%; 3, >30%) and staining intensity (0, none or weak staining; 1, moderate staining; 2, strong staining).9 The 90th and 75th percentile scores were used as cutoff values to classify CRC samples into high- and low-expressed subgroups for WISP3 expression.
Statistical analysis
The difference of clinicopathological characteristics between high- and low-expressed subgroups was evaluated by chi-square test. Difference in survival between high- and low-risk subgroups was compared using the Kaplan–Meier curve method and evaluated by log-rank test. Cox proportional hazards regression was used in multivariate model analysis. P<0.05 was considered as statistically significant. All the statistical analyses were performed using GraphPad Prism 5.0 software (Graphpad Software, San Diego, CA, USA).
Results
Here, we used a previously established COPA strategy to identify genes that display substantial expression changes in subpopulations of tumors from TCGA mRNA expression dataset. Top 20 outlier genes of 90th and 75th percentile were listed in Figure 1A. Among them WISP3 displays the highest COPA score (5.548), demonstrating a typical outlier model. As seen in Figure 1B, 21 and 58 cases were classified as WISP3 highly expressed based on the 90th and 75th percentile cutoffs, respectively; while no WISP3 outlier overexpression was seen in normal colorectal tissues (Figure 1B). We then performed meta-COPA on the expression model of WISP3 in other three independent CRC cohorts, as seen in Figure 1C–E, WISP3 ranked the top 1% of the outlier genes in all the three CRC microarray datasets. Together with our finding in TCGA CRC dataset, these findings further confirmed that WISP3 demonstrated an outlier expression model in CRC tumors.
We next analyzed the relationship between WISP3 outlier expression with clinical characteristics and survival. As seen in Table 1, WISP3 outlier expression was significantly associated with less aggressive phenotype such as small tumor size (90th outlier, P=0.0029), negative distant metastasis (90th outlier, P=0.0054), and early stage (75th outlier, P=0.0412) except that WISP2 75th outlier expression was significantly associated with positive lymph node metastasis (P=0.0365). Survival data analysis revealed that CRC patients without WISP3 outlier expression displayed a shorter overall survival time compared with those with WISP3 expression, but failed to reach a statistical significance (Figure 2).
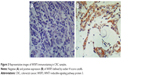
We further experimentally evaluated the clinical and prognostic significance of WISP3 in CRC patients by immunohistochemistry. Our immunohistochemical results showed that WISP3 was positively stained in the cytoplasm of cancer cells, occasionally positive in the stroma (Figure 3). If using moderate to strong staining in 10% of cancer cells as a cutoff, 55 cases (29.7%) were classified as positive staining, while no positive staining was seen in normal colorectal tissues. We then classified CRC patients according to 90th and 75th H score cutoff, and confirmed that positive WISP3 was more frequently observed in small tumors (T1–T2) than in large tumors (T2–T3) (Table 2). Furthermore, the survival data analysis found that 75th WISP3 outlier expression predicts better prognosis, and retained to be significant in multivariate analysis independent of other clinical prognostic factors such as grade, tumor size, and local and distant metastasis (Figure 4, Table 3).
Discussion
The strategy to compare the different expressed genes between cancerous and normal samples can identify general tumor biomarkers, but will not work for finding those genes where only expressed in a subset of cancer samples. The established outlier gene analysis methods have correctly prioritized oncogene and drug target ERBB2 in the breast cancer subsets.6 In this study, using COPA strategy, for the first time, we identified WISP3, displaying an outlier expression model across several large CRC cohorts, which has been further validated experimentally by immunohistochemistry. From the Oncomine database, WISP3 cannot be identified to be differently expressed between CRC and normal tissues. In a previous study, Davies et al10 demonstrated that WISP3 transcript levels showed no significant differences between CRC and normal colorectal tissues. The findings together with our results support that WISP3 is specifically expressed only in a subset of CRC patients.
We next evaluated the clinical implication of WISP3-positive CRC. Both the microarray data and immunohistochemical results indicated that WISP3 overexpressed CRC patients tended to display a relatively less aggressive phenotype. Our experimental data also suggest that WISP3 might be an independent survival factor for CRC. This finding is helpful to provide additional prognostic information to the current pathological stage system.
Downregulation or loss of function mutation of WISP3 gene was detected in various cancers, including aggressive breast cancers, lung cancers, and gastrointestinal carcinomas.11–13 In particular, WISP3 demonstrated a close association with inflammation-related cancers. Kleer et al11 found that WISP3 is specifically lost in inflammatory breast cancer (IBC) when compared to non-IBC, and loss of WISP3 expression contributes greatly to the aggressive phenotype of IBC. Kleer et al14 proposed that WISP3 and RhoC guanosine triphosphatase may act in concert to determine the highly aggressive IBC phenotype. The findings that WISP3 re-expression restores the differentiated epithelial phenotype in the breast and inhibits tumor cell motility and invasion in vitro and tumor growth in vivo, support its role as a tumor suppressor gene.15–17 Moreover, WISP3 is also shown to inhibit tumor-induced angiogenesis and causes epithelial-mesenchymal transition of breast epithelial cells.11,17 Mechanically, WISP3 exerts its tumor-suppressive function by several possible mechanisms. For example, Huang et al18 found that WISP3 can modulate E-cadherin expression in the breast epithelium through induction of snail and zinc finger E-box binding homeobox 1. Pal et al16 indicated that WISP3 acts to limit breast cancer invasion and metastasis by modulating bone morphogenetic proteins signaling via the Smad-independent TAK1/p38 pathway. Lorenzatti et al17 suggest that WISP3 decreases zinc finger E-box binding homeobox 1-mediated epithelial-mesenchymal transition and invasion in breast cancer by attenuation of insulin-like growth factor 1 receptor signaling. Therefore, the relationship with better prognosis identified in this study also suggests that WISP3 may also act as a candidate tumor suppressor gene for CRC, however, the exact mechanism remains to be clarified.
Recently, WISP3 has been regarded as a more selective, rational therapeutic target for cancers.19,20 Considering the close relationship between WISP3 knockdown and TAK1 activation, WISP3 deficient cancers may represent TAK1-dependent cancer subtypes.16,19 TAK1 inhibition has been shown to suppress tumor progression in preclinical models. Therefore, we proposed that CRC tumors with loss or downregulation of WISP3 expression may benefit from TSL-kinase interacting protein 1 inhibitors.
Conclusion
This study identified WISP3 as an outlier gene overexpressed in a subset of CRC. We demonstrated that WISP3-positive subgroups have a relatively less aggressive phenotype, thus enabling the identification of the patient population with a better prognosis. And we also suggest the molecular mechanism and therapeutic significance of WISP3 in CRC deserve further investigation.
Acknowledgments
This study was supported by Zhejiang Natural Science Foundation (LY16H160040 and LY16H160041), Huzhou Key Projects of Public Welfare (2014GZ11), and Zhejiang Applied Research Projects of Public Welfare (2014C33245).
Disclosure
The authors report no conflicts of interest in this work.
References
Wu B. Cancer outlier differential gene expression detection. Biostatistics. 2007;8(3):566–575. | ||
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. | ||
Barresi V, Reggiani Bonetti L, Ieni A, Caruso RA, Tuccari G. Histological grading in colorectal cancer: new insights and perspectives. Histol Histopathol. 2015;30(9):1059–1067. | ||
Mori T, Ikeda DD, Fukushima T, Takenoshita S, Kochi H. NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle. 2011;10(19):3284–3299. | ||
Lovf M, Nome T, Bruun J, et al. A novel transcript, VNN1-AB, as a biomarker for colorectal cancer. Int J Cancer. 2014;135(9):2077–2084. | ||
Rhodes DR, Ateeq B, Cao Q, et al. AGTR1 overexpression defines a subset of breast cancer and confers sensitivity to losartan, an AGTR1 antagonist. Proc Natl Acad Sci U S A. 2009;106(25):10284–10289. | ||
Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. | ||
Vilar E, Bartnik CM, Stenzel SL, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71(7):2632–2642. | ||
Huang HJ, Neven P, Drijkoningen M, et al. Association between tumour characteristics and HER-2/neu by immunohistochemistry in 1362 women with primary operable breast cancer. J Clin Pathol. 2005;58(6):611–616. | ||
Davies SR, Davies ML, Sanders A, Parr C, Torkington J, Jiang WG. Differential expression of the CCN family member WISP-1, WISP-2 and WISP-3 in human colorectal cancer and the prognostic implications. Int J Oncol. 2010;36(5):1129–1136. | ||
Kleer CG, Zhang Y, Pan Q, et al. WISP3 is a novel tumor suppressor gene of inflammatory breast cancer. Oncogene. 2002;21(20):3172–3180. | ||
LV J, Zou Y, Zhang C, Mao Z. Expressions of Cyr61 and WISP-3 in non-small cell lung cancer and its clinical significance. Zhongguo Fei Ai Za Zhi. 2010;13(12):1122–1126. | ||
Thorstensen L, Holm R, Lothe RA, et al. WNT-inducible signaling pathway protein 3, WISP-3, is mutated in microsatellite unstable gastrointestinal carcinomas but not in endometrial carcinomas. Gastroenterology. 2003;124(1):270–271. | ||
Kleer CG, Zhang Y, Pan Q, et al. WISP3 and RhoC guanosine triphosphatase cooperate in the development of inflammatory breast cancer. Breast Cancer Res. 2004;6(1):R110–R115. | ||
Pal A, Huang W, Toy KA, Kleer CG. CCN6 knockdown disrupts acinar organization of breast cells in three-dimensional cultures through up-regulation of type III TGF-β receptor. Neoplasia. 2012;14(11):1067–1074. | ||
Pal A, Huang W, Li X, Toy KA, Nikolovska-Coleska Z, Kleer CG. CCN6 modulates BMP signaling via the Smad-independent TAK1/p38 pathway, acting to suppress metastasis of breast cancer. Cancer Res. 2012;72(18):4818–4828. | ||
Lorenzatti G, Huang W, Pal A, Cabanillas AM, Kleer CG. CCN6 (WISP3) decreases ZEB1-mediated EMT and invasion by attenuation of IGF-1 receptor signaling in breast cancer. J Cell Sci. 2011;124(Pt 10):1752–1758. | ||
Huang W, Zhang Y, Varambally S, et al. Inhibition of CCN6 (Wnt-1-induced signaling protein 3) down-regulates E-cadherin in the breast epithelium through induction of snail and ZEB1. Am J Pathol. 2008;172(4):893–904. | ||
Leask A. CCN6 (WISP3): a new anti-cancer therapy? J Cell Commun Signal. 2010;4(4):199–200. | ||
Leask A. CCN6: a novel method of aTAKing cancer. J Cell Commun Signal. 2013;7(2):161–162. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.