Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Variants in PRKCE and KLC1, Potential Regulators of Type I Psoriasis
Authors Xing J, Wang Y, Zhao X, Li J, Hou R, Niu X, Yin G, Li X, Zhang K
Received 21 April 2022
Accepted for publication 15 June 2022
Published 1 July 2022 Volume 2022:15 Pages 1237—1245
DOI https://doi.org/10.2147/CCID.S371719
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Jianxiao Xing,1 Ying Wang,1 Xincheng Zhao,1 Junqin Li,1 Ruixia Hou,1 Xuping Niu,1 Guohua Yin,1 Xinhua Li,1 Kaiming Zhang2
1Shanxi Key Laboratory of Stem Cell for Immunological Dermatosis, Taiyuan Central Hospital of Shanxi Medical University, Taiyuan, 030009, Shanxi Province, People’s Republic of China; 2Shanxi Key Laboratory of Stem Cell for Immunological Dermatosis, Taiyuan Central Hospital, Taiyuan, 030009, Shanxi Province, People’s Republic of China
Correspondence: Kaiming Zhang, Taiyuan Central Hospital, No, 5 Dong San Dao Xiang, Jiefang Road, Taiyuan, Shanxi Province, People’s Republic of China, Tel +86-0351-5656080, Email [email protected]
Purpose: Psoriasis is a multifactorial disease with a complex genetic predisposition. The pathophysiology of psoriasis is associated with genetic variants. To better characterize gene variants in psoriasis and identify the relationship between clinical characteristics and variant genes in its pathogenesis.
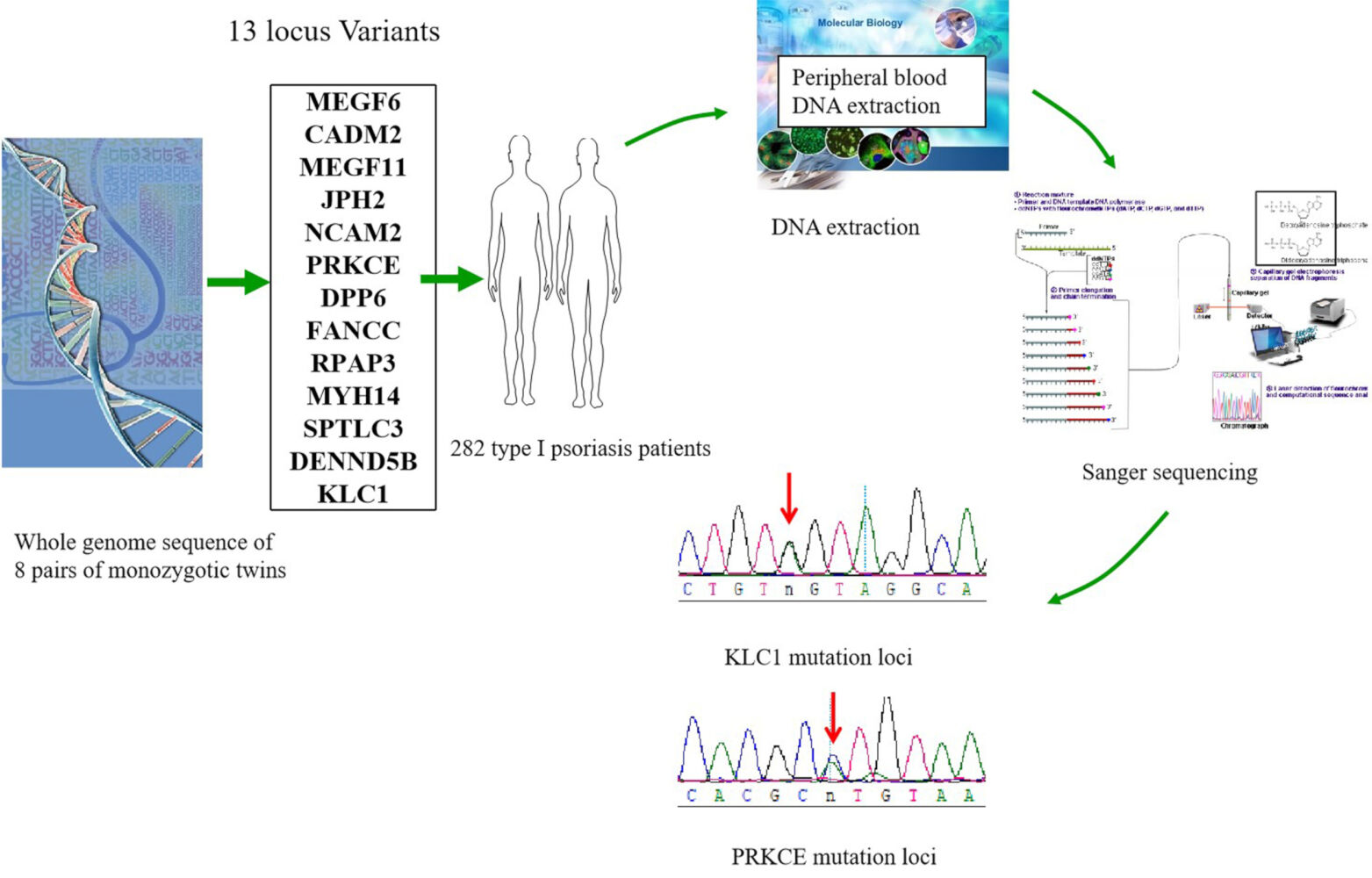
Patients and Methods: DNA was extracted and purified from eight pairs of monozygotic twins with psoriasis discordance and 282 type I psoriasis patients. Thirteen variable genes were amplified and sequenced using the Sanger method after whole genome sequencing.
Results: Thirteen genes were found to be variable in eight pairs of monozygotic twins with psoriasis discordance. Among the 13 genes, the variant frequencies of protein kinase C epsilon (PRKCE) (c.240T>C, 35.9% vs 47.7%, P < 0.05) and kinesin light chain 1 (KLC1) (c.216A>G, 2.9% vs 98.1%, P< 0.01) were significantly lower in psoriasis than in normal Asian individuals. Additionally, we found considerable differences in the relationship between variants in genes CADM2, JPH2, SPTLC3 and clinical characteristics stratified by medical history and family history. Moreover, the variants in MEGF6 (39.52% vs 22.50%, χ2=3.83, p < 0.05) showed a stronger association with the mild group (PASI ≤ 10) than the heavy group.
Conclusion: Our results provide a comprehensive correlation analysis of regulatory genes that are regulated in psoriasis. This integrated analysis offers novel insight into the pathogenic mechanisms involved in psoriasis.
Keywords: psoriasis, gene variants, PRKCE, KLC1
Graphical Abstract:
Introduction
Psoriasis is a common, genetic, chronic inflammatory skin disease that causes red, flaky patches on the skin.1,2 The worldwide prevalence of psoriasis is approximately 3.2%, or more than 7.4 million people, as estimated by Rachakonda et al.3 Psoriasis has a high recurrence rate and is considered a complex disease due to environmental and genetic factors.4
The genetic component is partly explained by its association with certain susceptibility loci.5 Understanding the genetic background of psoriasis is important for understanding6 the functional and structural property of the skin, identifying clinical biomarkers, discovering novel drug targets, and accelerating the journey towards personalized medicine.7,8 At present, more than 70 susceptibility genes are associated with psoriasis.9 Although most studies have focused on psoriasis susceptibility genes, others have shown that certain genetic variants play a regulatory role in the pathogenesis of psoriasis.10 The strongest susceptibility locus lies within the major histocompatibility complex (MHC).11 Studies have identified more than 60 disease susceptibility regions, highlighting the pathogenic involvement of genes related to Th17 cell activation. Moreover,12 genetic variants in the IL10 gene cluster were associated with psoriasis.
Because monozygotic (MZ) twins carry the theoretically same genetic information, they are more conducive to the study of susceptibility genes than sporadic populations.13 Recently, we obtained the whole genome sequences of eight pairs of monozygotic twins with psoriasis discordance and found 13 loci that were significantly different between normal individuals and patients with psoriasis in this paper. According to the onset age, psoriasis vulgaris was classified as type I (early onset, onset age <40) and type II (late onset, onset age ≥40). Type I psoriasis is strongly associated with onset age,16 anxiety, depression,17 HLA CW6, and18 ZFP36L1 mutations, etc.14,15 Many scholars suggested a genetic influence upon its age at onset, especially in type I psoriasis. To further investigate the contribution of these variants, we additionally examined these gene variants in 282 type I psoriatic patients and analyzed the relation between variants and clinical manifestation based on the monozygotic twins investigation.
Materials and Methods
Samples
Eight pairs of Chinese monozygotic (MZ) twins discordant in psoriasis with at least 5 years of medical history and their parents were enrolled with consent at Taiyuan Central Hospital.
In total, 282 patients diagnosed with vulgaris psoriasis were recruited from the outpatient center of the Taiyuan Central Hospital. All patients included in this study were of Han Chinese descent. Individuals with psoriasis were diagnosed by at least two dermatologists based on clinical and histopathological manifestations, and their clinical information was collected through a comprehensive clinical check-up by professional investigators. Self-reported information from a standard questionnaire was used to collect demographic and other characteristics (severity, medical history, and family history) from the patients and to exclude any other systemic, infectious, autoimmune, atopic, or malignant disease and to determine whether they received systemic treatment in the 6 months prior to collection. None of the patients had hypertension, gout, asthma or cafe au lait spot. All participants provided written informed consent, including parents or legal guardian of participants under 18 years of age provided informed consent. The study protocol was approved by the ethics committee of Taiyuan Central Hospital.
Characteristics of the Subjects
The clinical features of MZ twins samples (4 pairs of women and 4 pairs of men, range from 11 to 51 years with the mean age 30 years) are shown in Supplemental Table 1.
The clinical characteristics of the study subjects are shown in Supplemental Table 2. In this study, we investigated 282 Chinese Han individuals with type I psoriasis. Their age of onset is less than 40 and pertain to early onset, which all belong to type I patients with psoriasis.
Whole Genome Sequencing and Variant Identification
Each qualified sample of genomic DNA from whole blood was sequenced on the Illumina HiSeq platform using paired-end reads according to the manufacturer’s instructions.19 The following types of reads were removed (1) containing a sequencing adapter; (2) low-quality base >50%; and (3) “N” base >10%. All clean data from each sample were mapped to the human reference genome HG19 by Burrows Wheeler Aligner software (BWA, v0.75). Duplicate reads were marked by Picard tools.20 We used 3 different methods to call SNPs and InDels and get overlap of them to ensure the accuracy of variant calling SAMTOOLS mpileup,21 recommended best practices for variant analysis with the Genome Analysis Toolkit (GATK) and Freebayes. We identified the DNM discordance between co-twins by checking the concordance genotype between them. We genotyped our DNMs with Sanger sequencing to confirm our DNM detection accuracy.
DNA Extraction
Genomic DNA was extracted from the peripheral whole blood of the 282 patients using a Blood Genomic DNA Midi Kit (Cwbio Biotech, Beijing, China) according to the manufacturer’s instructions. All DNA samples were dissolved in water and stored at −20°C until use.
Sanger Sequencing
Sequencing primers (Supplemental Table 3) were designed for the 13 single-nucleotide polymorphisms (SNPs) using Primer Premier 5.0. Genomic DNA was amplified using the Bio-Rad PCR System. Thermal cycling was performed as follows: 5 min at 96°C for 10 cycles (20 seconds at 96°C; 30 seconds for touchdown at 52–62°C; and 60 seconds at 72°C), followed by 35 cycles (20 seconds at 96°C; 30 seconds at 52°C; and 60 seconds at 72°C), and ending with 5 min at 72°C.
Statistical Analysis
Information on the variants frequency in 8624 normal Asian individuals was obtained from the Pubvar database (https//www.pubvar.com/). Amplicons were bidirectionally sequenced using an ABI 3730 system. We performed sequence analysis by using Variants Surveyor software. Variants included hybrid variants and homozygous variants. The variation frequency was calculated with the following equation (hybrid+ homozygous ×2)/307/2. Differences in patient demographics (eg, stage, sex, age, severity, medical history, and family history) were evaluated with SPSS version 18.0. The chi-square ( 2) test was used to test the relationship between psoriasis and the investigated factors. Statistical significance was set at P < 0.05.
2) test was used to test the relationship between psoriasis and the investigated factors. Statistical significance was set at P < 0.05.
Results
Thirteen Variant Loci Were Filtered in the Monozygotic Twins
In this study, blood sample from eight pairs of monozygotic twins with psoriasis discordance was analyzed by whole genome sequencing. We obtained 13 variants loci, such as kinesin light chain 1 (KLC1, c.216A>G), protein kinase C epsilon (PRKCE, c.240T>C), multiple epidermal growth factor-like domains 6 (MEGF6, c.96 A>G), cell adhesion molecule 2 (CADM2, c.263 A>G), multiple epidermal growth factor-like domains 11 (MEGF11, c.80 T>G), neural cell adhesion molecule 2 (NCAM2, c.236 G>T), dipeptidyl peptidase like 6 (DPP6, c.174 G>T), myosin heavy chain 14 (MYH14, c.461 T>C), serine palmitoyltransferase long-chain base subunit 3 (SPTLC3, c.481 T>C), FA complementation group C (FANCC, c.134 C>T), DENN domain containing 5B (DENND5B, c.96 A>G) junctophilin 2 (JPH2, c.195 T>G) and RNA polymerase II associated protein 3 (RPAP3, c.146 C>G) (Table 1). Interestingly, all of the 13 above mentioned variant loci were found only in the normal homozygote, but none of them in psoriatic homozygote. Variants exist in normal populations, indicating their potential prevention to psoriasis.
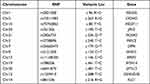 |
Table 1 Thirteen Mutant Loci in eight Pairs of Monozygotic Twins |
The Variation Frequency of PRKCE and KLC1 Were Lower Than Normal Individuals
When analyzing the clinical information of monozygotic twins, it shows that 75% of the patients are under the age of 40, that is, they are classified as Type I according to the type. Therefore, we speculate that the above 13 mutations are closely related to Type I. We screened 282 cases of Type I psoriasis for further validation. Among 614 loci in 282 psoriasis patients, 240 loci in protein kinase C epsilon (PRKCE) were mutated (c.240T>C), and the variation frequency in patients with psoriasis (36.3%) was significantly lower than that in normal Asian individuals (47.7%, P<0.05). Moreover, 216 loci in kinesin light chain 1 (KLC1) were mutated (c.216A>G), and the variation frequency in patients with psoriasis (1.2%) was significantly lower than that in normal Asian individuals (98.1%, P<0.01). In addition, the mean variation frequencies of the genes MEGF6 (20.2% vs 25.9%), CADM2 (71.4% vs 86.1%), MEGF11 (59.2% vs 61.8%), NCAM2 (32.4% vs 33.6%), DPP6 (81.2% vs 83.9%), MYH14 (17.7% vs 22.0%), SPTLC3 (64.5% vs 66.1%), FANCC (4.4% vs 4.60%) and DENND5B (0.53% vs 0.6%) were lower in patients with psoriasis than in normal individuals, though all of P values were higher than 0.05. However, the variation frequencies of JPH2 (86.1% vs 85.12%) and RPAP3 (28.0% vs 26.9%) were higher in patients with psoriasis than in normal individuals (Figure 1).
 |
Figure 1 Differences in variation frequencies between psoriasis patients and normal individuals. *P<0.05, **P<0.01. |
Occurrence of Psoriasis and Its Relationship with the Sociodemographic Characteristics of Participants
Medical history, the PASI score and family history were used to clarify the different clinical subtypes. Therefore, the relationship between medical history, the PASI score, family history and variant genes was investigated. Regarding medical history, PASI and family history, 282 patients were grouped, respectively. The medical history group was divided into two ranks less than 20 years (58.2%) and more than 20 years (41.8%). CADM2 (χ2 = 9.29, P<0.05), JPH2 (χ2 = 8.47, P<0.05), SPTLC3 (χ2 = 20.65, P<0.01) was significantly different between patients with different medical histories (Figure 2).
Patients were selected based on disease severity, which was assessed using PASI score. The number of patients in the mild group (PASI score ≤10) was 209 (74.1%), and the number of patients in the moderate-to-severe group (PASI score >10) was 73 (25.9%). Analysis of the PASI score revealed a significant association with variation in MEGF6 (χ2 = 6.06, P<0.05).
Patients were also grouped according to family history positive (27.3%) and negative (49.3%). Next, we further analyzed the relationship between family history and variant genes and found that CADM2 (χ2 = 6.08, P<0.05), JPH2 (χ2 = 10.98, P<0.01), SPTLC3 (χ2 = 8.51, P<0.01) was significantly associated with family history, suggesting that the variation in CADM2, JPH2, SPTLC3 may be the risk factor for psoriasis in individuals who have a family member with psoriasis.
Univariate and Multivariate Analysis of Family History on Disease Characteristics
For research purposes, we rearranged the population data and divided individuals with psoriasis into two groups, as shown in Table 2. Research has shown that with the extension of time, new mutations will emerge and old ones will be repaired. Therefore, to eliminate the impact of time on mutation gene, we divided into two groups based on medical history and then compare different PASI and family history of patients. In the under 20 years medical history group and over 20-year medical history group, we analyzed the relationship between gender, age, PASI, family history and gene mutation. For the comparison of family history of the two groups, we found that CADM2 (χ2 =5.89, 94.87% vs 76.56%, p<0.05) was a risk factor for having family history with type I psoriasis. Similarly, when compared to patients with under 20 years medical history group and over 20 year medical history group, there were differences in MEGF6 (χ2 =3.86, 39.52% vs 22.5%, p<0.05) according to PASI of type I psoriasis.
 |
Table 2 Chi-Square Test the PASI and Family History in Patients with Different Medical Histories |
Discussion
Based on screening studies and genome-wide association studies (GWASs),22,23 a large number of susceptibility genes that are involved in the pathogenesis of complex diseases, such as cancer, psoriasis, and other major human diseases, have been identified. Concerning psoriasis,24,25 more than 70 susceptible genes have been identified in the last 30 years. In our study, we use next-generation sequencing technique to assess the status of 13 genes, and we finally found that PRKCE and KLC1 had mutated in psoriasis patients. The results of the study indicated that genetic variation has a very close correlation with clinical features.26
A pathogenic role for PRKCE has been found in several types of cancers, as it is able to promote proliferation and inhibit apoptosis.27–29 Constitutively active PRKCE has been found in small cell lung cancer and epithelial cells of the colon associated with tumor formation.30 The PRKCE acts as a regulator of PLD activity, and this inhibition is mediated by its regulatory domain.31 Moreover, the novel partial loss-of-function defect in PRKCE impairs AKT activation via compromised mTORC2 complex function.32 According to studies of patients with psoriasis, the PI3K/AKT pathway is an important signaling pathway that regulates the hyperproliferation of keratinocytes.33 It has also been reported that a variation at the phosphorylation site of KLC1, Ser517/Ser520, affects AMP-activated protein kinase (AMPK) to suppress low glucose concentrations and block granule movement.34 In this study, we demonstrated that the variation rates of PRKCE and KLC1 were significantly lower in psoriasis patients than in normal Asian individuals, consistent with the results reported by Salo-Mullen EE, indicating that the described PRKCE and KLC1 variants may be a regulatory mechanism for the onset of psoriasis.
Medical history and the PASI score are commonly used in clinical trials on psoriasis, and35 this scoring algorithm greatly expands options for quantifying treatment outcomes in cost-effectiveness analyses of psoriasis therapies. To further evaluate the relationship between mutant genes and clinical characteristics, we grouped patients with psoriasis according to sex, age, medical history, PASI score and family history. The variation rate was distinctly different according to medical history, PASI score and family history. We found that the CADM2, JPH2 and SPTLC3 variation have correlation with more than 20 years of medical history in individuals with type I psoriasis. Moreover, moderate-to-severe (PASI score >10) were more commonly observed in individuals with a MEGF6 variation.36,37 Indeed, several studies have shown the significant impact of the onset of psoriasis not only in patients with a medical history and a high PASI score but also in their family and close relatives.38 The literature supports some differences between familial and sporadic cases of psoriasis.39 Differences in the strength of the association between the mutant gene and family history have been reported for the MHC, including a strong association of HLA-C*06 and HLA-B*27 with psoriasis.
Similarly, in our study, we found that the variation frequency of CADM2, JPH2 and SPTLC3 was higher in the familial group than in the sporadic group, and many studies also show that they have a strong relationship with psoriasis.40 Cell adhesion molecules (CADMs) immunoglobulin super family is involved in the maintenance of cell adhesion, polarity and tumor suppression. Moreover,41 the CADM2/Akt pathway is involved in the inhibitory effect of miR-21-5p downregulation on proliferation and apoptosis.42 JPH2 mutation caused perturbations in intracellular calcium signaling and marked cardiomyocyte hyperplasia.43 SPTLC3 is associated with disorders such as cardiovascular and metabolic diseases, while44 psoriasis severity and the prevalence of metabolic syndrome observed a dose–response relationship.45 Studies have reported that genetic factors interact with molecular pathways involving TNF-α, IL-23/IL-17 axis. This provides the basis for treatment options for type I psoriasis.
Limitations
In addition, our study has some limitations. More works are needed to verify our findings and illustrate the detailed mechanism of these mutation genes based on larger sample size in the future.
Conclusion
In summary, we demonstrated that the variation rates of KLC1 and PRKCE were significantly lower in psoriasis than normal, indicating these two genes could serve as potential novel regulatory genes for psoriasis. We also showed considerable differences in the relationship between clinical characteristics and mutant genes. Our findings are important for the promotion of large panels in patient populations with psoriasis and for clinical genetic testing for patient management.
Ethics Statement
The study was approved by the institutional ethics committee of Taiyuan City Central Hospital (2016005). The study was carried out with the informed consent of the patients themselves and in accordance with the Declaration of Helsinki Principles.
Acknowledgments
The authors are grateful for funding from the National Natural Science Foundation of China (nos. 81773336, 81602768, and 81803146). The funding contributed to the design of the study and the collection, analysis, and interpretation of data. We would like to thank AJE (https//www.aje.cn/) for English language editing.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Lin Y, Liu L, Sheng Y, et al. A catalog of potential putative functional variants in psoriasis genome-wide association regions. PLoS One. 2018;13:e0196635. doi:10.1371/journal.pone.0196635
2. Rachakonda TD, Dhillon JS, Florek AG, Armstrong AW. Effect of tonsillectomy on psoriasis a systematic review. J Am Acad Dermatol. 2015;72:261–275. doi:10.1016/j.jaad.2014.10.013
3. Tang L, Cheng Y, Zhu C, et al. Integrative methylome and transcriptome analysis to dissect key biological pathways for psoriasis in Chinese Han population. J Dermatol Sci. 2018;91:285–291. doi:10.1016/j.jdermsci.2018.06.001
4. Ammar M, Bouchlaka-Souissi C, Soumaya K, et al. Failure to find evidence for deletion of LCE3C and LCE3B genes at PSORS4 contributing to psoriasis susceptibility in Tunisian families. Pathol Biol. 2014;62:34–37. doi:10.1016/j.patbio.2013.10.003
5. Ogawa K, Okada Y. The current landscape of psoriasis genetics in 2020. J Dermatol Sci. 2020;99(1):2–8. doi:10.1016/j.jdermsci.2020.05.008
6. Caputo V, Strafella C, Termine A, et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J Cell Mol Med. 2020;24(23):13554–13563. doi:10.1111/jcmm.15742
7. Wang J, Li W, Shi Y, et al. Germline mutation landscape of Chinese patients with familial breast/ovarian cancer in a panel of 22 susceptibility genes. Cancer Med. 2019;8:2074–2084. doi:10.1002/cam4.2093
8. Rahman P, Elder JT. Genetics of psoriasis and psoriatic arthritis a report from the GRAPPA 2010 annual meeting. J Rheumatol. 2012;39:431–433. doi:10.3899/jrheum.111242
9. Fan X, Wang H, Sun L, et al. Fine mapping and subphenotyping implicates ADRA1B gene variants in psoriasis susceptibility in a Chinese population. Epigenomics. 2019;11:455–467. doi:10.2217/epi-2018-0131
10. Dębniak T, Soczawa E, Boer M, et al. Common variants of ZNF750, RPTOR and TRAF3IP2 genes and psoriasis risk. Arch Dermatol Res. 2014;306:231–238. doi:10.1007/s00403-013-1407-9
11. Capon F. The genetic basis of psoriasis. Int J Mol Sci. 2017;18(12):2526. doi:10.3390/ijms18122526
12. Galimova E, Rätsep R, Traks T, et al. Interleukin-10 family cytokines pathway: genetic variants and psoriasis. Br J Dermatol. 2017;176(6):1577–1587. doi:10.1111/bjd.15363
13. Li J, Lin H, Hou R, et al. Multi-omics study in monozygotic twins confirm the contribution of de novo mutation to psoriasis. J Autoimmun. 2020;106:102349. doi:10.1016/j.jaut.2019.102349
14. Hüffmeier U, Lascorz J, Becker T, et al. Characterisation of psoriasis susceptibility locus 6 (PSORS6) in patients with early onset psoriasis and evidence for interaction with PSORS1. J Med Genet. 2009;46:736–744. doi:10.1136/jmg.2008.065029
15. Sun LD, Cheng H, Wang ZX, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–1009. doi:10.1038/ng.690
16. Remröd C, Sjöström K, Svensson A. Psychological differences between early- and late-onset psoriasis: a study of personality traits, anxiety and depression in psoriasis. Br J Dermatol. 2013;169(2):344–350. doi:10.1111/bjd.12371
17. Fatema F, Ghoshal L, Saha A, et al. Early-onset versus late-onset psoriasis: a comparative study of clinical variables, comorbidities, and association with HLA CW6 in a tertiary care center. Indian J Dermatol. 2021;66(6):705. doi:10.4103/ijd.ijd_45_21
18. Wang H, Wang C, Wang Z, et al. Identification of ZFP36L1 as an early-onset psoriasis risk gene demonstrates opposite associations with leprosy and psoriasis in the Chinese population. J Eur Acad Dermatol Venereol. 2020;34(9):e520–e523. doi:10.1111/jdv.16437
19. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi:10.1126/science.1109557
20. Zhao Y, Forst CV, Sayegh CE, et al. Molecular and genetic inflammation networks in major human diseases. Mol Biosyst. 2016;12:2318–2341. doi:10.1039/C6MB00240D
21. Liu QP, Wu LS, Li FF, et al. The association between GJB2 gene polymorphism and psoriasis a verification study. Arch Dermatol Res. 2012;304:769–772. doi:10.1007/s00403-012-1273-x
22. Choon SE, Lai NM, Mohammad NA, et al. Clinical profile, morbidity, and outcome of adult-onset generalized pustular psoriasis: analysis of 102 cases seen in a tertiary hospital in Johor, Malaysia. Int J Dermatol. 2014;53:676–684. doi:10.1111/ijd.12070
23. Chen H, Toh TK, Szeverenyi I, et al. Association of skin barrier genes within the PSORS4 locus is enriched in Singaporean Chinese with early-onset psoriasis. J Invest Dermatol. 2009;129:606–614. doi:10.1038/jid.2008.273
24. Stuart PE, Nair RP, Ellinghaus E, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42:1000–1004. doi:10.1038/ng.693
25. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi:10.1101/gr.107524.110
26. Jain K, Basu A. The multifunctional protein kinase C-epsilon in cancer development and progression. Cancers. 2014;6:860–878. doi:10.3390/cancers6020860
27. Baxter G, Oto E, Daniel-Issakani S, et al. Constitutive presence of a catalytic fragment of protein kinase C epsilon in a small cell lung carcinoma cell line. J Biol Chem. 1992;267:1910–1917. doi:10.1016/S0021-9258(18)46033-4
28. Perletti GP, Folini M, Lin HC, et al. Overexpression of protein kinase C epsilon is oncogenic in rat colonic epithelial cells. Oncogene. 1996;12:847–854.
29. Gorin MA, Pan Q. Pan protein kinase C epsilon an oncogene and emerging tumor biomarker. Mol Cancer. 2009;8:9. doi:10.1186/1476-4598-8-9
30. Pan Y, Yuan C, Cheng C, et al. Frequency and clinical significance of NF1 mutation in lung adenocarcinomas from East Asian patients. Int J Cancer. 2019;144:290–296. doi:10.1002/ijc.31871
31. Alcantara D, Elmslie F, Tetreault M, et al. SHORT syndrome due to a novel de novo mutation in PRKCE (Protein Kinase Cvarepsilon) impairing TORC2-dependent AKT activation. Hum Mol Genet. 2017;26:3713–3721. doi:10.1093/hmg/ddx256
32. Zhang M, Zhang X. The role of PI3K/AKT/FOXO signaling in psoriasis. Arch Dermatol Res. 2019;31:183–191.
33. McDonald A, Fogarty S, Leclerc I, et al. Cell-wide analysis of secretory granule dynamics in three dimensions in living pancreatic beta-cells evidence against a role for AMPK-dependent phosphorylation of KLC1 at Ser517/Ser520 in glucose-stimulated insulin granule movement. Biochem Soc Trans. 2010;38:205–208. doi:10.1042/BST0380205
34. Wason JM, Dudbridge F. A general framework for two-stage analysis of genome-wide association studies and its application to case-control studies. Am J Hum Genet. 2012;90:760–773. doi:10.1016/j.ajhg.2012.03.007
35. Chandran V, Schentag CT, Brockbank JE, et al. Familial aggregation of psoriatic arthritis. Ann Rheum Dis. 2009;68:664–667. doi:10.1136/ard.2008.089367
36. Raposo I, Carvalho C, Bettencourt A, et al. Psoriasis pharmacogenetics HLA-Cw*0602 as a marker of therapeutic response to ustekinumab. Eur J Dermatol. 2017;27:528–530. doi:10.1684/ejd.2017.3071
37. Makondi PT, Lee CH, Huang CY, et al. Prediction of novel target genes and pathways involved in bevacizumab-resistant colorectal cancer. PLoS One. 2018;13:e0189582. doi:10.1371/journal.pone.0189582
38. O’Rielly DD, Jani M, Rahman P, et al. The genetics of psoriasis and psoriatic arthritis. J Rheumatol Suppl. 2019;95:46–50. doi:10.3899/jrheum.190119
39. Bejaoui Y, Witte M, Abdelhady M, et al. Genome-wide association study of psoriasis in an Egyptian population. Exp Dermatol. 2017;28:623–627. doi:10.1111/exd.13926
40. Liu N, Yang C, Bai W, et al. CADM2 inhibits human glioma proliferation, migration and invasion. Oncol Rep. 2019;41(4):2273–2280. doi:10.3892/or.2019.7010
41. Li X, Chen D, Li M, et al. The CADM2/Akt pathway is involved in the inhibitory effect of miR-21-5p downregulation on proliferation and apoptosis in esophageal squamous cell carcinoma cells. Chem Biol Interact. 2018;288:76–82. doi:10.1016/j.cbi.2018.04.021
42. Landstrom AP, Weisleder N, Batalden KB, et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42(6):1026–1035. doi:10.1016/j.yjmcc.2007.04.006
43. Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–141. doi:10.1038/ng.507
44. Singh S, Young P, Armstrong AW. Relationship between psoriasis and metabolic syndrome: a systematic review. G Ital Dermatol Venereolhttps. 2016;151(6):663–677.
45. Campione E, Cosio T, Di Prete M, et al. Experimental pharmacological management of psoriasis. J Exp Pharmacol. 2021;13:725–737. doi:10.2147/JEP.S265632
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

