Back to Journals » Clinical Ophthalmology » Volume 17
Variability of Replicates of Intraocular Inflammatory Biomarkers in Ocular Fluid Samples Analyzed with Multiplex Assays
Authors Lamoureux D , Wong DT, Felfeli T
Received 5 May 2023
Accepted for publication 20 July 2023
Published 8 September 2023 Volume 2023:17 Pages 2653—2663
DOI https://doi.org/10.2147/OPTH.S417821
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Daniel Lamoureux,1 David T Wong,2,3 Tina Felfeli2,4
1Northern Ontario School of Medicine University, Thunder Bay, ON, Canada; 2Department of Ophthalmology and Vision Sciences, University of Toronto, Toronto, ON, Canada; 3Department of Ophthalmology, St. Michael’s Hospital, Unity Health Toronto, Toronto, ON, Canada; 4The Institute of Health Policy, Management and Evaluation, University of Toronto, Toronto, ON, Canada
Correspondence: Tina Felfeli, University Health Network, Toronto General Hospital, Eaton Building, 10th Floor, Toronto, ON M5G 2C4, Canada, Tel +1 647 678 1634, Fax +1 416 340 3459, Email [email protected]
Purpose: Certain factors such as instrumental and sample processing errors may contribute to variability of ocular biofluid samples when they are run as replicates with multiplex assays. There is a paucity of literature on the variability of replicates in multiplex assays. This study aims to evaluate whether there is significant variability in replicate analyses of multiplex assays.
Methods: A total of 152 human ocular biofluid samples (51 aqueous humor and 101 vitreous) were collected and assayed for 27 cytokine biomarker concentrations (pg/mL). Samples were evaluated as replicates (duplicate analysis) at four different time points. Statistical methods including paired samples t-test, 3-way ANOVA, intraclass correlation coefficient (ICC; < 0.5– 0.75=poor-moderate, 0.75-> 0.90 =good-excellent reliability), and coefficients of variation (CV) were employed to evaluate for statistical significance, with Bonferroni corrected P=0.002.
Results: Among the 4104 biomarker replicate assays for aqueous humor and vitreous, two analytes (PDGF-BB and IL-7) had a statistically significant difference between the sampled concentrations of the replicates in vitreous samples (mean (diff)=2.05, P< 0.001, mean (diff)=1.56, P< 0.001, respectively). Majority of the ICC values fell within the good-excellent range (86% of samples) with a minority falling in the poor-moderate range (14% of samples). More variability was noted in the vitreous humour, with five analytes (IL-2, IL-10, IL-12(p70), IL-13, IL-17) demonstrating an average ICC of less than 0.5. The CV calculated for each set of replicates suggested that 93% of replicates had an acceptable level of quantitative assay variability (CV< 20%).
Conclusion: This study demonstrates that the analysis of most biomarkers in ocular fluids may not require the use of replicates. However, certain analytes such as PDGF-BB and IL-7 may require the use of replicates to ensure reliable results. Caution should be taken when applying these findings to other laboratory settings as our study was conducted by an experienced technician using a standardized protocol. In less standardized settings, replicates may be required in order to ensure accuracy of results. These findings may guide researchers with the design of their studies on ophthalmic biomarker analysis.
Keywords: cytokines, aqueous, vitreous, biomarkers, assay
Introduction
Cytokine biomarkers have unlocked new avenues to explore disease processes and may be utilized to assess for disease severity and prognostication.1–5 With the advent of multiplex assays, researchers are able to evaluate multiple cytokines simultaneously, leading to a greater understanding of the pathophysiology of a disease, as well as the discovery of novel therapeutic targets.6 Ocular fluid samples analyzed using multiplex assays can be analyzed as singles or as replicates (duplicates or triplicates). Samples analyzed as singles are evaluated once in their entirety, while replicates refer to division of samples for repeated analysis, which allows for averaging of concentrations computed. The benefit of running the assays in replicates versus singles may be error minimization and increasing reliability in case of potential outliers. The disadvantages of analyzing samples as replicates are reduced sample volume availability for assay analysis, an increase in reagent consumption, and additional time and cost related to multiple multiplex kits and analyses required.7
Sampling of intraocular fluids can pose various challenges; the ocular repositories in which the fluid is collected, such as the aqueous humor and the vitreous humor, can only accommodate the removal of a finite volume of fluid and carries associated procedure risks. Furthermore, it can be challenging to evaluate these samples with multiplex assays, particularly with the minimal volumes associated with replicates. There is a paucity of literature on the variability of ocular biofluid samples when they are run as replicates with multiplex assays. It is important to ensure the accuracy of biomarker analysis as these results may have a direct impact on the patient’s management, and ultimately, their health outcomes. The use of biomarker analysis with artificial intelligence has been demonstrated in many ophthalmic diseases, such as glaucoma, uveitis, uveal melanoma, age-related macular degeneration, corneal and ocular surface diseases, and retinal occlusive diseases.8–12
Herein, we evaluated the variability of intraocular inflammatory biomarkers in multiplex assays of intraocular biofluid samples.
Materials and Methods
Study Design
This is an experimental study. Samples from eyes of patients undergoing eye surgery were used for this study. Exclusion criteria included an age less than 18 years, a previous vitrectomy of the eye in question, and dense vitreous hemorrhage at the time of initial assessment for recruitment. This study was performed in accordance with the Declaration of Helsinki and Health Insurance Portability and Accountability Act,13 and was approved by the Unity Health Toronto (REB# 20-142), and University of Toronto (RIS# 40230) Research Ethics Boards.
Sample Collection and Handling
Samples collected included 15 aqueous humor specimens and 20 vitreous specimens (see Figure 1), each with a volume ranging from 0.1mL to 0.3mL. Samples were undiluted and immediately aliquoted to yield 152 human ocular fluid aliquots. Vitreous aliquots contained approximately 0.075mL per tube (four tubes per sample), while aqueous sample volume was divided into two to three tubes depending on the sample volume. The samples were collected during cataract surgery, pars plana vitrectomy and scleral buckle surgical procedures. Aqueous humor samples were taken from the anterior chamber at the beginning of the procedure, while vitreous samples were obtained following the insertion of the trocars and before starting the vitrectomy procedure in order to avoid dilution of the samples. The samples were immediately placed on ice, then stored at 4°C for 5 hrs followed by storage in a freezer with a temperature of −80°C until time of assay analysis. All of the samples were collected during a 1-week timeframe. The same sample analyses including replicates for each of the analytes (duplicate analysis) were completed at four different time points (analysis 1, analysis 2, analysis 3, analysis 4) following sample collection. This methodology and assay results have been utilized by the authors in other works.14
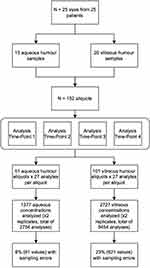 |
Figure 1 Flowchart demonstrating data collection and analysis process. |
Sample Analysis
All ocular samples were assayed using the Bio-Plex Pro-Human Cytokine Grp I Panel 27-Plex 96-well kit (Cat# M500KCAF0Y, Bio-Rad, United States) assay (pg/mL), as per the manufacturer’s instructions. All analyses were done on the same kits with the same lot number for the Bio-Rad multiplex assay. The aqueous and vitreous samples were diluted with 1:2 and 1:1 concentration of the standard diluent buffer, respectively. Each well contained 50μL of fluorescently dyed magnetic microspheres diluted in an assay buffer and 50μL of diluted aqueous humor or vitreous samples, standards or blanks. All samples were centrifuged prior to setting up the assays. Bio-Plex Manager Software was utilized to assess the concentrations. Sampling errors were determined by this software, including the “low bead number” error and the “region selection” error. The software can detect a “low bead number” error if it detects a total number of region events collected less than or equal to one-fourth of 100 (or number of events selected by the user) as well as a total number of events collected less than or equal than three times 100 (number of analytes selected times the events selected by the user). The “region selection” error is triggered when the software looks to see if a selected region has zero beads in it. This could indicate that the wrong region was chosen or that the beads did not go to the region. Values with sampling errors were included in the dataset in order to capture the true variability of the assay results.
The 27 analytes that were examined for expression include interleukin- (IL-)1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17, basic fibroblast growth factor (FGF), eotaxin, granulocyte-colony stimulating factor (G-CSF), granulocyte macrophage-colony stimulating factor (GM-CSF), interferon gamma (IFN-γ), interferon gamma-induced protein-10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein 1 (MIP-1)α, MIP-1β, platelet-derived growth factor- (PDGF-) BB, regulated on activation, normal T cell expressed and secreted (RANTES), tumor necrosis factor alpha (TNF-α), and vascular endothelial growth factor (VEGF). These analytes are from a pre-defined set of cytokines from the manufacturer, and they are based on compatibility and feasibility. The assay kit has also been utilized in other projects to assess cytokines in intraocular fluids.15
Primary and Secondary Outcomes
The main outcome of this study was to assess the variability of replicate analyses of intraocular inflammatory biomarkers in multiplex assays of intraocular biofluid samples. We assessed this by evaluating the CV as well as the ICC between replicate analysis of each analyte from the same sample. Further, for secondary outcomes, we aimed to determine if other factors such as ocular fluid type and analyte have an impact on the CV of replicate analyses.
Statistical Analysis
Descriptive statistics were used to determine general trends. For each biomarker in their respective ocular medium (aqueous humor, vitreous humor), statistical significance was evaluated within replicates by conducting a paired samples t-test. Concentrations from each time point (1 week, 3 months, 9 months, and 15 months) were compiled for these comparisons. The original p-value was set at 0.05. A Bonferroni correction factor was applied to evaluate the statistical significance. As there are 27 analytes, a Bonferroni correction factor of 27 was used, resulting in a Bonferroni corrected P value of 0.002.
The coefficient of variation (CV) for all sets of replicates was calculated, by dividing the standard deviation by the mean for each difference in the replicate sets (by analyte) in their respective ocular medium (aqueous and vitreous). The CV is a unitless value that demonstrates the relative dispersion of a dataset, allowing the comparison of datasets with means that greatly differ from one another. A CV value of less than 20% was deemed acceptable based on previously published reports.16,17 Given that previous studies have implemented a CV threshold of 25% for their evaluation,18,19 a sensitivity analysis was conducted to compare how this threshold would impact the values in this study. Further, we determined how many CV values were greater or equal to an ideal CV threshold of 5%.
Moreover, a three-way ANOVA was utilized to examine the effects of analysis time-point, ocular fluid type, and analyte on the CV. Finally, the intraclass correlation coefficient (ICC) was calculated to evaluate for similarity of replicate concentrations for each analyte and time point. An ICC of <0.5–0.75 was considered to be poor-moderate and 0.75->0.90 was deemed good-excellent reliability.20
All statistical analyses were performed with the use of STATA software (version 17.0; Stata Corporation, College Station, TX, USA).
Results
A total of 4104 biomarker replicate assays for aqueous humor (1377) and vitreous (2727) were included in the analysis (see Figure 1). All samples were obtained from 25 eyes of 25 patients. Patient demographics can be found in Appendix A. The smallest mean concentration in the aqueous humor and vitreous humor was associated with IL-13, with a concentration of 12.4 ± 17.4 pg/mL and 8.2 ± 1.4, respectively (Table 1). Similarly, the largest mean concentration in the aqueous humor and vitreous humor was associated with MCP-1, with a concentration of 5509.1 ± 3808.6 pg/mL and 6944.9 ± 4622.6, respectively. The range of individual concentrations for all analytes ranged from 5.5 pg/mL to 16,729 pg/mL. The most common diagnosis from the samples was rhegmatogenous retinal detachment with 20% of samples representing this diagnosis. Further, 56% of samples were from females and 44% from males, with a mean age of 63 years.
 |
Table 1 Mean Concentration and Standard Deviation for All Analytes in Aqueous Humor and Vitreous Humor |
Two analytes (PDGF-BB and IL-7) had a statistically significant difference between the sampled concentrations of the replicates in vitreous samples (mean (diff)=2.05, P<0.001, mean (diff)=1.56, P<0.001, respectively). Table 2 includes a summary of the analysis for replicate analyses for aqueous and vitreous samples. For the CV, 268 of the 4104 (6.5% of samples) biomarker replicate assays had a CV greater or equal to 20%, with 164 of the 4104 (4.0% of samples) replicate assays having a CV greater or equal to 25%, and 1976 of the 4104 (48.1% of samples) having a CV greater or equal to 5%. From the 268 replicate sets with a CV greater or equal to 20%, 49 (18%) were of aqueous humor, and 219 (82%) were of vitreous humor. Figure 2 demonstrates the CV results for each analyte in both the aqueous and vitreous, while Figures 3 and 4 demonstrate the CV results for each analyte with their respective ocular mediums.
 |
Table 2 Variability of Replicates for Cytokine Biomarkers in Aqueous and Vitreous Humor Samples |
 |
Figure 2 Coefficient of variation of all analytes for all samples (aqueous, vitreous). The horizontal dashed line represents the acceptable coefficient of variation limit (20%). |
 |
Figure 3 Coefficient of variation of all analytes (aqueous humor). The horizontal dashed line represents the acceptable coefficient of variation limit (20%). |
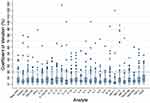 |
Figure 4 Coefficient of variation of all analytes (vitreous humor). The horizontal dashed line represents the acceptable coefficient of variation limit (20%). |
The ICC calculations yielded 185 of 216 values (86%) in the good to excellent range (ICC ≥ 0.75), and 31 of 216 values (14%) in the poor to moderate range (ICC < 0.75). Table 2 includes a summary of the ICC results. Five of the analytes analyzed in the vitreous humor (IL-2, IL-10, IL-12(p70), IL-13, IL-17) had an average ICC less than 0.50 indicating poor reliability. Detailed analysis of ICC results for each of the four analysis time-points is included in Appendix B. In the three-way interaction for evaluating the effects of analysis time-point, ocular fluid type, and analyte on the CV, there was no statistically significant difference (F(78, 3888) = 0.32, p=1.000).
Among all analyses, 6% (81) aqueous humor values and 23% (621) vitreous humor values contained sampling errors . There were a total of 54 sampling errors detected by the assay analysis software, in which 36 were marked as “low bead number” errors, and 18 were marked as “region selection” errors. These sampling errors were specific for each multiplex assay run, and common among all analytes. That is, each analyte for the 1 week multiplex assay run contained the same sampling errors for each sample. Similarly, this was the case for the 3 months run, the 9 months run and the 15 months run. Of the “low bead number” sampling errors, 18 of 36 (50%) were unique to one replicate, while the remaining 18 (50%) sampling errors were common to a set of replicates. This differed from the “region selection” sampling error, which had 14 of 18 (78%) errors unique to one replicate and remaining 4 of 18 (22%) were common to a set of replicates.
Discussion
The purpose of this study was to examine the variability of replicate analyses of intraocular biomarkers in both aqueous and vitreous humors, using multiplex assays. Studies have demonstrated that cytokine assay results may be impacted by factors including but not limited to storage temperature, time period between obtaining the sample and storing it, and repeated freeze-thawing.14,21–23 Other factors inherent to the multiplex assay kit may contribute to variability of assay results, along with instrumental error associated with the multiplex assay procedure. Our findings demonstrated generally good consistency in replicate analyses of analytes within aqueous and vitreous humors in the same multiplex assay. However, there were some notable variability amongst specific analytes analysed in vitreous samples. Notably, two analytes (PDGF-BB and IL-7) had a statistically significant difference between the sampled concentrations of the replicates in vitreous samples. Additionally, certain analytes of the interleukin family (IL-2, IL-10, IL-12(p70), IL-13, IL-17) demonstrated poor reliability in the vitreous humor, with an average ICC of less than 0.5. In general, most of the ICC values (86% of samples) fell within the good-excellent range, demonstrating good reliability for the majority of replicate analyses.
Depending on the application, a CV value of less than 5% is ideal, although an acceptable limit of 20% is often utilized in research applications.24 The majority of biomarker replicate assays in this study had a CV less than 20% (93.5% of replicate assays), with 96.0% having a CV less than 25% and 51.9% having an ideal CV of less than 5%. With the majority of biomarker replicate assays having a CV less than the acceptable limit of 20% and the majority of the ICC values falling in the good-excellent range (86% of samples), the use of singles appears to be favourable and appropriate for certain research applications. This methodology is further supported when considering the aforementioned disadvantages of analyzing samples as replicates, such as reduced sample volume available for assay analysis, an increase in reagent consumption, and additional time and costs associated with the assay procedure. However, our results suggest that specific analytes from vitreous samples, such as PDGF-BB and IL-7, may be more susceptible to replicate variability in multiplex assays than others. IL-2, IL-10, IL-12(p70), IL-13 and IL-17 also demonstrated poor reliability in vitreous humor assays with an average ICC less than 0.50. This would have important implications in studies that evaluate these specific analytes in vitreous humor. In these cases, it may be more suitable to evaluate samples in replicates, to ensure reliable results. It is difficult to hypothesize why these analytes had more variability than the others; however, we suspect it is a culmination of a few different factors. Notably, the expression levels of most of these analytes were on the lower end of the spectrum when considering mean concentration levels. Additionally, it is known that certain cytokine concentrations may be more susceptible to storage temperatures along with freeze/thaw cycles.25 We believe this might be attributed to molecular features specific to the cytokine. Additionally, it is known that the medium in which a cytokine is stored may have an impact on its concentration.25 Thus, we hypothesize that there may be a component in the reagent solution, which increased the variability of the assay results. Similarly, Chaturvedi et al found that assay performance decreased with an increasing number of markers on the panel and attributed this to a likely interference from other markers having an impact on the measured marker.17 Finally, additional factors such as user error and environmental conditions could have also contributed to some of the inconsistencies.
A CV threshold of 20% was deemed appropriate for this study; however, it is important to note that this threshold should vary depending on the specific application and goals of the analysis. Our sensitivity analysis of an increased CV threshold of 25% demonstrated that 6.5% of biomarker replicate assays had high variability, as compared to 4% of cases with high variability associated with a 20% CV cut-off value. Studies have demonstrated the potential of inflammatory biomarker analysis, for diagnostics and clinical decision-making.26,27 In these types of applications, a CV threshold of 20% may be inappropriate and clinical applications utilizing ophthalmic biomarker analysis must adjust their statistical parameters accordingly.
Similarly, the acceptability for sampling errors will be dependent on the specific application in question. In our study, the proportion of sampling errors was relatively small for samples from aqueous humor, as compared to those from vitreous humor (6% and 23%, respectively). This may be due to the difference in viscosities between the aqueous and vitreous humors, which may limit the ability of the assay beads to mix evenly, leading to inconsistencies.28 Additionally, with 23 of the 27 analytes having a lower mean concentration in the vitreous humor as compared to the aqueous humor, there may be an increased probability of having sampling errors in the vitreous since these generally had lower mean concentrations in our study. A study evaluating reproducibility and correlations of multiplex cytokine levels in serum found suboptimal reproducibility of a cytokine assay at low or sub-pg/mL concentrations, while other studies testing at higher cytokine levels, maximized reliability.29 The lower mean concentrations in the vitreous humor may be attributed to the vitreous humor having a larger compartment than the aqueous humor and thus, more fluid available to dilute the analytes. Furthermore, half of all sampling errors were common to replicate sets, with the other half being unique to one replicate in the replicate set. This is noteworthy as singles that contain sampling errors will require the discarding of a complete sample, whereas a sample in which one replicate contains a sampling error may simply require the discarding of the replicate containing the sampling error. This would allow for the concentration of the replicate without the sampling error to be salvaged, a scenario that is not ideal, albeit one that is better than the entire sample being rendered obsolete. Moreover, a portion of the variability in the cytokine concentrations between ocular samples can be attributed to the number of various diseases included in our study. With various disease processes inducing the release of different cytokines, there will surely be a difference between cytokine concentrations between samples. For future studies, there may be value to analyze the samples with the grouping of disease processes. Due to our small sample size of diseases, we did not do this for the current study.
The strengths of this study include a relatively large dataset and a large number of aliquots for a study of this nature. Furthermore, the samples and procedures were carried out in a single center, ensuring a uniform collection and handling process and ensuring that the same instruments are used throughout the study. The specimens were stored immediately, and analyzed at multiple time points in order to increase the sample size. As a way of ensuring comparability of biomarker concentrations, all analyses were done on the same kits with the same lot number. The generalizability of our findings, however, as a result may be limited to Bio-Rad multiplex assay kits. Future studies may evaluate the variability in replicate analyses with other assays to ensure consistency of findings.
Conclusion
In conclusion, this study demonstrates that the analysis of most biomarkers in ocular fluids may not require the use of replicates. The analysis of certain analytes such as PDGF-BB and IL-7 may require the use of replicates to ensure reliable results. Caution should be taken when applying these findings to other laboratory settings as our study was conducted by an experienced technician using a standardized protocol. In less standardized settings, replicates may be required in order to ensure accuracy of results. These findings are important in helping to guide the methodology for future studies involving the use of multiplex assays and inflammatory biomarkers in ophthalmic fluid samples.
Acknowledgments
The authors would like to thank Professor Bruce Weaver (Lakehead University, Northern Ontario School of Medicine University) for his contributions and guidance with the statistical analyses carried out in this study.
Disclosure
Dr David T Wong reports grants and personal fees from Bayer, Roche, Novartis; personal fees from Alcon, AbbVie, Apellis, Ripple Therapeutics, Zeiss, and Bausch Health; equity from ArcticDX; and on the board of directors for 20/20 Innovation Hub, outside the submitted work. The authors report no other conflicts of interest in this work. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
1. Dorgham K, Quentric P, Gökkaya M., et al. Distinct cytokine profiles associated with COVID-19 severity and mortality. J Allergy Clin Immunol. 2021;147(6):2098–2107. doi:10.1016/j.jaci.2021.03.047
2. Thanou A, Jupe E, Purushothaman M, Niewold TB, Munroe ME. Clinical disease activity and flare in SLE: current concepts and novel biomarkers. J Autoimmun. 2021;119:102615. doi:10.1016/j.jaut.2021.102615
3. Hao X, Yi C, Wang Y, et al. Identification of intraocular inflammatory mediators in patients with endophthalmitis. Mol Vis. 2016;22:563–574.
4. Chen H, Zhang X, Liao N, Wen F. Assessment of biomarkers using multiplex assays in aqueous humor of patients with diabetic retinopathy. BMC Ophthalmol. 2017;17(1):176. doi:10.1186/s12886-017-0572-6
5. Usui Y, Tsubota K, Agawa T, et al. Aqueous immune mediators in malignant uveal melanomas in comparison to benign pigmented intraocular tumors. Graefes Arch Clin Exp Ophthalmol. 2017;255(2):393–399. doi:10.1007/s00417-016-3541-5
6. Weinstein JE, Pepple KL. Cytokines in uveitis. Curr Opin Ophthalmol. 2018;29(3):267–274. doi:10.1097/ICU.0000000000000466
7. Raggatt PR. Duplicates or singletons?—An analysis of the need for replication in immunoassay and a computer program to calculate the distribution of outliers, error rate and the precision profile from assay duplicates. Ann Clin Biochem. 1989;26(1):26–37. doi:10.1177/000456328902600104
8. Pucchio A, Krance SH, Pur DR, Miranda RN, Felfeli T. Artificial intelligence analysis of biofluid markers in age-related macular degeneration: a systematic review. Clin Ophthalmol. 2022;16:2463–2476. doi:10.2147/OPTH.S377262
9. Pur DR, Krance SH, Pucchio A, Miranda RN, Felfeli T. Current uses of artificial intelligence in the analysis of biofluid markers involved in corneal and ocular surface diseases: a systematic review. Eye (Lond). 2022. doi:10.1038/s41433-022-02307-9
10. Pur DR, Krance S, Pucchio A, Bassi A, Miranda RN, Felfeli T. Emerging applications of bioinformatics and artificial intelligence in the analysis of biofluid markers involved in retinal occlusive diseases: a systematic review. Graefes Arch Clin Exp Ophthalmol. 2023;261(2):317–336. doi:10.1007/s00417-022-05769-5
11. Bassi A, Krance SH, Pucchio A, Pur DR, Miranda RN, Felfeli T. The application of artificial intelligence in the analysis of biomarkers for diagnosis and management of uveitis and uveal melanoma: a systematic review. Clin Ophthalmol. 2022;16:2895–2908. doi:10.2147/OPTH.S377358
12. Pucchio A, Krance S, Pur DR, Bassi A, Miranda R, Felfeli T. The role of artificial intelligence in analysis of biofluid markers for diagnosis and management of glaucoma: a systematic review. Eur J Ophthalmol. 2022;11206721221140948. doi:10.1177/11206721221140948
13. World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi:10.1001/jama.2013.281053
14. Felfeli T, Nestor B, Park J, Wong DT. Effect of sample collection and storage on biological stability of cytokines in human aqueous humor and vitreous samples. Invest Ophthalmol Vis Sci. 2022;63(7):2370–A0054.
15. Ke D, Hong Y, Jiang X, Sun X. Clinical features and vitreous biomarkers of early-onset type 2 Diabetes mellitus complicated with proliferative diabetic retinopathy. Diabetes Metab Syndr Obes. 2022;15:1293–1303. doi:10.2147/DMSO.S362074
16. Reed GF, Lynn F, Meade BD. Use of coefficient of variation in assessing variability of quantitative assays. Clin Vaccine Immunol. 2002;9(6):1235–1239. doi:10.1128/CDLI.9.6.1235-1239.2002
17. Chaturvedi AK, Kemp TJ, Pfeiffer RM, et al. Evaluation of multiplexed cytokine and inflammation marker measurements: a methodologic study. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1902–1911. doi:10.1158/1055-9965.EPI-11-0221
18. Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and mesoscale discovery, for human cytokine profiling. J Immunol Methods. 2009;340(1):55–64. doi:10.1016/j.jim.2008.10.002
19. Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56(2):314–318. doi:10.1373/clinchem.2009.135087
20. Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15(2):155–163. doi:10.1016/j.jcm.2016.02.012
21. Aziz N, Nishanian P, Mitsuyasu R, Detels R, Fahey JL. Variables that affect assays for plasma cytokines and soluble activation markers. Clin Diagn Lab Immunol. 1999;6(1):89–95. doi:10.1128/CDLI.6.1.89-95.1999
22. Balaiya S, Grover S, Murthy RK, Chalam KV. Freezing adversely affects measurement of vascular endothelial growth factor levels in human aqueous samples. Clin Ophthalmol. 2011;5:81–85. doi:10.2147/OPTH.S15837
23. Flower L, Ahuja RH, Humphries SE, Mohamed-Ali V. Effects of sample handling on the stability of interleukin 6, tumour necrosis factor-alpha and leptin. Cytokine. 2000;12(11):1712–1716. doi:10.1006/cyto.2000.0764
24. Tworoger SS, Hankinson SE. Use of biomarkers in epidemiologic studies: minimizing the influence of measurement error in the study design and analysis. Cancer Causes Control. 2006;17(7):889–899. doi:10.1007/s10552-006-0035-5
25. Simpson S, Kaislasuo J, Guller S, Pal L. Thermal stability of cytokines: a review. Cytokine. 2020;125:154829. doi:10.1016/j.cyto.2019.154829
26. Inami W, Shibuya M, Kumagai T, Makita J, Shinoda K. A case of intraocular lymphoma diagnosed by subretinal fluid biopsy. Int Med Case Rep J. 2022;15:111–115. doi:10.2147/IMCRJ.S345149
27. Velez G, Roybal CN, Colgan D, Tsang SH, Bassuk AG, Mahajan VB. Precision medicine: personalized proteomics for the diagnosis and treatment of idiopathic inflammatory disease. JAMA Ophthalmol. 2016;134(4):444–448. doi:10.1001/jamaophthalmol.2015.5934
28. Le Guezennec X, Lye M. Improving bead-based multiplexing in bodily fluids. Genet Eng Biotechnol News. 2016;36(2):12–13. doi:10.1089/gen.36.02.07
29. Wong HL, Pfeiffer RM, Fears TR, Vermeulen R, Ji S, Rabkin CS. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3450–3456. doi:10.1158/1055-9965.EPI-08-0311
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
