Back to Journals » Clinical Epidemiology » Volume 8
Validity of the coding for herpes simplex encephalitis in the Danish National Patient Registry
Authors Jørgensen LK, Dalgaard L, Østergaard LJ , Andersen NS , Nørgaard M , Mogensen TH
Received 16 January 2016
Accepted for publication 3 March 2016
Published 31 May 2016 Volume 2016:8 Pages 133—140
DOI https://doi.org/10.2147/CLEP.S104379
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Henrik Sørensen
Laura Krogh Jørgensen,1 Lars Skov Dalgaard,1 Lars Jørgen Østergaard,1 Nanna Skaarup Andersen,2 Mette Nørgaard,3 Trine Hyrup Mogensen1
1Department of Infectious Diseases, Aarhus University Hospital, Aarhus, 2Department of Clinical Microbiology, Odense University Hospital, Odense, 3Department of Clinical Epidemiology, Aarhus University Hospital, Aarhus, Denmark
Background: Large health care databases are a valuable source of infectious disease epidemiology if diagnoses are valid. The aim of this study was to investigate the accuracy of the recorded diagnosis coding of herpes simplex encephalitis (HSE) in the Danish National Patient Registry (DNPR).
Methods: The DNPR was used to identify all hospitalized patients, aged ≥15 years, with a first-time diagnosis of HSE according to the International Classification of Diseases, tenth revision (ICD-10), from 2004 to 2014. To validate the coding of HSE, we collected data from the Danish Microbiology Database, from departments of clinical microbiology, and from patient medical records. Cases were classified as confirmed, probable, or no evidence of HSE. We estimated the positive predictive value (PPV) of the HSE diagnosis coding stratified by diagnosis type, study period, and department type. Furthermore, we estimated the proportion of HSE cases coded with nonspecific ICD-10 codes of viral encephalitis and also the sensitivity of the HSE diagnosis coding.
Results: We were able to validate 398 (94.3%) of the 422 HSE diagnoses identified via the DNPR. Hereof, 202 (50.8%) were classified as confirmed cases and 29 (7.3%) as probable cases providing an overall PPV of 58.0% (95% confidence interval [CI]: 53.0–62.9). For “Encephalitis due to herpes simplex virus” (ICD-10 code B00.4), the PPV was 56.6% (95% CI: 51.1–62.0). Similarly, the PPV for “Meningoencephalitis due to herpes simplex virus” (ICD-10 code B00.4A) was 56.8% (95% CI: 39.5–72.9). “Herpes viral encephalitis” (ICD-10 code G05.1E) had a PPV of 75.9% (95% CI: 56.5–89.7), thereby representing the highest PPV. The estimated sensitivity was 95.5%.
Conclusion: The PPVs of the ICD-10 diagnosis coding for adult HSE in the DNPR were relatively low. Hence, the DNPR should be used with caution when studying patients with encephalitis caused by herpes simplex virus.
Keywords: HSE, viral encephalitis, positive predictive value, validation studies, DNPR, sensitivity
Introduction
Herpes simplex encephalitis (HSE) is the most common form of sporadic encephalitis worldwide and remains a disease with high mortality and long-term morbidity.1,2 If untreated, the mortality reaches 70%, and remains ˜20% in the case of appropriate intravenous acyclovir treatment.3,4 At least 50% of HSE survivors suffer from neurological deficits such as aphasia and amnesia.5 The incidence of HSE is approximately two to four cases per million population per year.4 In Sweden, a population-based estimate of 2.2 cases per million population per year was calculated in a nationwide study.6 It is expected that 90% of the cases are caused by herpes simplex virus type 1 (HSV-1), and ˜10% are caused by HSV type 2 (HSV-2), the latter more often being associated with aseptic meningitis.7 Thus, HSE remains a disease requiring continuous investigation to better understand the epidemiology, pathogenesis, and determinants of prognosis.
Population-based health care databases, such as those developed in Denmark, may constitute a cost-effective way of conducting epidemiological studies on HSE patients. The large size of the databases offers a potential for precise estimates even when studying rare outcomes or exposures. Typically, the data are collected for administrative purposes, and the risk of recall bias and nonresponse bias is therefore minimized.8 However, researchers who conduct observational studies using existing data are unable to control the data collection and quality. Therefore, the utility of these databases relies on the validity of the registered data.
To our knowledge, no previous studies have thoroughly validated HSE diagnoses in hospital registries. Hence, the aim of this study was to assess the positive predictive value (PPV) of the HSE diagnosis coding at patient discharge in the population-based Danish National Patient Registry (DNPR) using data from medical record reviews and microbiological data as a reference standard. Furthermore, we wished to estimate the sensitivity of the HSE diagnosis coding.
Methods
Data sources
The Danish National Health Service provides universal and unrestricted tax-supported health care. This facilitates free access to hospitals for all Danish residents. Since 1968, a unique ten-digit civil registry number (Civil Personal Registration [CPR] number) has been assigned to all Danish residents by the Danish Civil Registry System.9 The CPR number allows accurate and highly valid individual-level record linkage of data between Danish registries.10 Our study relied upon linkage of data from the DNPR, the Danish Microbiology Database (MiBa), departments of clinical microbiology, and patient medical records.
Danish National Patient Registry
The DNPR contains data from all admissions to Danish somatic hospitals since 1977, and all visits to emergency rooms and outpatient clinics since 1994. The registry contains administrative information such as dates of admission and discharge, place of admission, and primary and secondary diagnosis codes. The diagnoses are coded at patient discharge by medical doctors using the Danish version of the International Classification of Diseases, eighth revision (ICD-8) (1977–1993), and ICD, tenth revision (ICD-10) (since 1994). The diagnoses are subsequently entered in the hospital registry by a medical secretary and transmitted electronically to the DNPR. DNPR data are continuously updated with complete nationwide coverage since 1978.11
Danish Microbiology Database
The MiBa is a nationwide, automatically updated database of microbiological test results. Since January 2010, all microbiological test reports from the departments of clinical microbiology have been transferred electronically to MiBa.12
Patient medical records
Information from patient medical records includes record notes, radiology descriptions, and results from microbiological and biochemical tests.
Identification of coded HSE cases
We searched the DNPR for all patients who received a primary or secondary diagnosis code indicating HSE (ICD-10 codes B00.4 “Herpesviral [herpes simplex] encephalitis” and B00.4A “Herpesviral [herpes simplex] meningoencephalitis”) during the period of 2004–2014. The majority of infectious diseases are coded in the ICD-10 system chapters A00–B99, whereas diseases of the nervous system are coded in the chapters G00–G99. The corresponding code of B00.4 in the chapters for diseases of the nervous system is G05.1 “Encephalitis, myelitis, and encephalomyelitis in viral diseases classified elsewhere”. Accordingly, we additionally included the Danish subcode G05.1E “Herpesviral encephalitis” in our study. This code was, however, closed on January 1, 2012 in the updated Danish ICD-10 version and replaced by G051.U “Encephalitis in viral diseases classified elsewhere”.13
To ensure a uniform cohort, pediatric patients (<15 years of age) were excluded. Furthermore, only first-time HSE episodes were considered. For this reason, we excluded patients with a diagnosis code indicating HSE according to both the ICD-10 (B00.4, B00.4A, G05.1) and ICD-8 classifications (“Meningoencephalitis ex herpete simplici” – 054.03) recorded in the DNPR before the HSE admissions identified during 2004–2014. This was possible by utilizing the complete diagnostic history of these patients obtained from the DNPR dating back to 1977.
Microbiological data and medical record validation
To validate the specific diagnoses of HSE, we collected data on the patients identified in the DNPR from MiBa, two departments of clinical microbiology, and patient medical records linked by their CPR numbers. Confirmed HSE was primarily defined as a positive finding of HSV-1 DNA by polymerase chain reaction (PCR) techniques of the cerebrospinal fluid (CSF) samples and/or intrathecal HSV-1 antibody production (by enzyme-linked immunosorbent assay [ELISA]). The microbiological data on the patients diagnosed in the period of 2010–2014 were assessed using MiBa. As far as possible, a confirmed microbiological diagnosis of all the patients was sought. Therefore, microbiological test results of a limited number of patients diagnosed before 2010 were obtained from two of the seven microbiological departments in Denmark.
We retrieved and reviewed medical records of all the remaining patients identified in the DNPR – that is, those diagnosed before 2010 – and patients whose diagnosis could neither be confirmed nor be disproved by microbiological findings. All medical records were reviewed by one of the authors (LKJ). Available data including doctor’s notes, written radiology reports, and results from laboratory tests were assessed. In the majority of the cases, the information available was identical with the information available to the physician at time of discharge, but in some cases, diagnostic information was received shortly after the hospital discharge.
If the microbiological findings demonstrated HSV-2 or HSV type not specified (HSV-NS) by PCR of the CSF and/or intrathecal HSV-2/HSV-NS antibody production, the patients had to fulfill at least two of the following additional criteria to be considered a confirmed case: 1) acute signs of parenchymatous brain dysfunction (focal neurological deficits, decreased consciousness, and/or seizures), 2) positive brain imaging findings (computed tomography [CT]/magnetic resonance imaging [MRI]) of inflammation in frontotemporal lobes and/or electroencephalogram findings suggestive of encephalitis, and or 3) pleocytosis in CSF (≥5 white blood cells/mm3). Patients who according to their medical record did not undergo lumbar puncture or had negative microbiological HSV findings, but still fulfilled at least two of the aforementioned other criteria, were considered probable cases of HSE. The aforementioned inclusion criteria for confirmed and probable HSE were based on previously established definitions from Granerod et al14 and Persson et al.15 If the criteria were not met, or if symptoms were explained by another disease, patients were not considered cases of HSE.
Consequently, the cases were classified as “confirmed HSE”, “probable HSE”, or “no evidence of HSE”. All cases with an uncertain diagnosis based on the information available were discussed among the authors (LKJ and THM), and decisions on confirmation or exclusion of these cases were made according to consensus agreement. In cases with double-positive microbiological findings of both varicellazoster virus (VZV) and HSV, the medical history was evaluated, and the cases with additional dermatomal zoster rash were excluded, but otherwise, they were considered cases of HSE.
Sensitivity analysis
A full evaluation of the sensitivity based on the data sources available was not possible. However, to estimate the sensitivity of the aforementioned HSE diagnoses, we searched the DNPR for hospitalized patients aged ≥15 years, who received nonspecific encephalitis ICD-10 diagnosis codes from January 1, 2010 to December 31, 2014. The following ICD-10 codes were used: A86.9 “Viral encephalitis without specification”, G04.9 “Encephalitis, myelitis and encephalomyelitis, without specification”, G04.9A “Encephalitis without specification”, and G05.1U “Encephalitis in viral diseases classified elsewhere”. Both primary and secondary diagnoses were included. If patients subsequently received a HSE-specific diagnosis code, they were excluded from this analysis.
To validate the nonspecific diagnoses of viral encephalitis, we assessed microbiological data from MiBa of these patients. In the analysis regarding coding sensitivity, the patients were considered a case of HSE if the MiBa data demonstrated positive finding of HSV-1 DNA by PCR of the CSF samples or intrathecal HSV-1 antibody production.
Statistical analyses
The study outcome was the PPV of HSE diagnoses, defined as the proportion of patients with a diagnosis of HSE in the DNPR, who had definite and probable HSE according to their medical records and microbiological data. The PPV was calculated for patients registered by each of the three ICD-10 codes indicating HSE and for the whole study population. We stratified the PPVs by type of diagnosis (primary and secondary), as we expected the PPV to be markedly higher for HSE coded as the primary cause of the admission. Furthermore, we stratified the PPV by period (2004–2007, 2008–2011, 2012–2014) to investigate whether the validity differed over time and by type of department (infectious diseases and neurology, as opposed to other departments). For each PPV, the corresponding 95% confidence interval (CI) was estimated using the method for binomial proportions.16 A chi-square test for linear trend in PPVs over the study period was made.
The sensitivity was estimated as the identified confirmed cases with HSE-specific diagnosis codes during 2010–2014 divided by the same cases added with the identified HSE cases coded with nonspecific ICD-10 diagnosis codes for viral encephalitis. The corresponding 95% CI was estimated.
We analyzed the data using Stata Software (v 13.1; Stata Corp., College Station, TX, USA). Clinical data from medical records were systematically collected using EpiData Software (v 2.0.3.15; EpiData Association, Odense, Denmark). The study was approved by the Danish Data Protection Agency (journal no 2007-58-0010, case no 1-16-02-581-14) and authorization to collect the relevant data from the medical files was provided by the National Board of Health (Journal no 3-3013-886/1). According to Danish law, no written informed consent is needed in studies based on registry data.
Results
Study population
In the DNPR, we identified 422 adult hospital-admitted patients with a diagnosis code indicating HSE during 2004–2014. None of the patients had previous episodes of HSE. The median age of the 422 patients was 60.1 years (interquartile range, 43.5–73.2). A total of 228 (54%) were female.
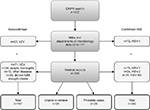
Figure 1 schematically shows the validation process of the HSE diagnoses. We were able to validate 398 (94.3%) of the 422 HSE diagnoses of adult patients identified via the DNPR. Of these, 95 (23.9%) of the diagnoses were defined by microbiological data from MiBa (n=77, 81.1%) and departments of microbiology (n=18, 18.9%). From the remaining 327 patients, we were able to retrieve medical records from 303 (92.7%). The 24 patients without available medical records were sporadically distributed in the different regions of Denmark. Of the 24 patients, 20.8% were diagnosed with HSE as a secondary diagnosis but had no other obvious similarities that could suggest that they represented a selected group of patients. The 24 patients were excluded before the analyses because of the missing information.
We classified 202 (50.8%) as being confirmed HSE cases and 29 (8.9%) as probable cases of HSE (Figure 1). Thus, 167 (42.0%) patients were not considered cases of HSE. The majority of these (n=94, 56.3%) were instead positive for VZV in the CSF. Thirty-four patients (20.4%) more likely had viral meningitis as they had no signs of parenchymatous brain dysfunction, nonetheless with positive HSV-2 (n=29) or HSV-NS (n=5) in the CSF. Ten patients (6%) were misclassified cases in the DNPR. Of these, eight patients had hepatic encephalopathy, and two patients had serological findings of Borrelia Burgdorferi and Legionella Pneumophila in their CSF. Additionally, 29 patients (17.4%) did not meet the inclusion criteria to be considered as probable cases of HSE. Of these 29 patients, the majority presented with clinical signs of parenchymatous brain dysfunction such as decreased consciousness or epileptic seizure but lacked positive brain imaging findings, and lumbar puncture was not performed. These cases may represent several possible diseases, that is, other types of viral encephalitis, limbic encephalitis, epilepsy, connective tissue diseases, stroke, etc. One patient had positive brain imaging showing temporal inflammation on the MRI but lacked other signs of parenchymatous brain dysfunction. Likewise, five patients had pleocytocis in the CSF but did not fulfill any additional criteria. Seven of the patients had a characteristic zoster rash but without microbiological findings of VZV in the CSF.
Validation of HSE diagnoses
The results from the validation of the HSE coding are shown in Table 1. The overall PPV of the three HSE diagnosis codes, including confirmed and probable cases, was 58.0% (95% CI: 53.0–62.9). When restricting the HSE definition to confirmed cases, the overall PPV of the HSE diagnoses in the DNPR was 50.8% (95% CI: 45.7–55.8). The coding validity did not differ significantly between the three different HSE diagnosis codes.
PPVs of HSE diagnosed in 2004–2007, 2008–2011, and 2012–2014 were 56.2% (95% CI: 47.5–64.7), 54.1% (95% CI: 46.3–61.8), and 68.1% (95% CI: 57.5–77.5), respectively. Although the PPV was highest in 2012–2014, the increase was not significant (P-value for trend =0.13). The PPVs stratified by type of diagnosis (primary or secondary) showed no major differences from the overall PPV. If the patient was diagnosed at a department of infectious diseases or a department of neurology, the PPV was 66.3% (95% CI: 60.1–72.1), which was significantly higher than the PPV of diagnoses from other departments (43.8%; 95% CI: 35.6–52.3).
A total of 635 patients received one of the aforementioned nonspecific diagnosis codes of encephalitis during 2010–2014. The majority received the diagnosis code “Viral encephalitis without specification” (A86.9) (n=341, 53.7%). Among these 635 patients, 27 had HSV in the CSF. Five patients had positive HSV-1, seven patients had positive HSV-2, and 15 patients had positive HSV-NS. Of note, 105 of the confirmed HSE patients (Figure 1) were diagnosed during 2010–2014. An approximated estimation of sensitivity is thereby 95.5% (95% CI: 89.7–98.5) when only including the HSV-1 positive. If all HSV-positive cases with a nonspecified ICD-10 code were true cases of HSE, the estimated sensitivity would be 79.5% (95% CI: 71.7–86.1).
Discussion
In this study, we examined the validity of the ICD-10 coding for HSE in the DNPR. We generally found very low PPVs. This result indicates a profound absence of agreement between DNPR coding (potentially covering encephalitis due to HSV), medical records, and microbiological data. We found no variation in the PPV stratified by study period and diagnosis type. Our results reveal a higher PPV if the patient was diagnosed at a specialized department of neurology or infectious diseases versus another medical department. Furthermore, we found a relatively high sensitivity of the ICD-10 HSE coding in the DNPR.
To our knowledge, there is no previous investigation of the validity of the HSE coding in the DNPR. The majority of previous studies validating ICD-10 diagnosis coding in the DNPR have found PPVs of ≥80% using both medical records and laboratory data as reference standard.17–20 Thereby, the PPVs found in this study were low as compared to PPV estimates for most other diagnoses. Recently, a systematic review investigated the data quality and research potential of the DNPR and reported substantial variation in the data validity with PPVs varying from 15% to 100%.11 This underscores the need of diagnosis validation before using data of the DNPR for research.
Attaining a validation rate of 94.3%, the results from this study can be considered generalizable to the Danish population. However, some limitations need to be considered in the interpretation of the results. First, we did not estimate the proportion of HSE patients not registered in the DNPR. Moreover, some HSE patients may have been registered with other diagnosis codes. Thus, full evaluation of the sensitivity, specificity, and negative predictive value was not possible. However, we did estimate the sensitivity based on identification of HSE patients coded with nonspecific ICD-10 codes for viral encephalitis. These results revealed a high sensitivity of the HSE-specific diagnosis codes. Furthermore, when the prevalence is low, the PPV is a good approximation of specificity.21 Second, only one reviewer (LKJ) evaluated most of the medical records, which might have reduced the internal validity of the study. However, the inclusion and exclusion criteria obtained from the medical records were quite precise and objective. Therefore, only few cases ended up with an uncertain diagnosis. These were evaluated and discussed by several of the authors. Third, we may have overestimated the PPV by including the probable cases. Fourth, 20% of the nonconfirmed cases had positive HSV-2 in the CSF but no signs of brain parenchymal involvement, and therefore did not fulfill the criteria of being cases of HSE. They were, instead, considered cases of viral meningitis rather than HSE. This decision may underestimate the PPVs of this study because cases of mild meningoencephalitis may have been excluded. Even if these cases were categorized as cases of HSE, the PPV would be <67%. Fifth, cross-reactions between VZV and HSV during the serological diagnostic laboratory procedures (ELISA) have been reported, which could result in an underestimation of the PPV.22 Finally, it is difficult to establish consensus among physicians regarding definitions of encephalitis, meningoencephalitis, and aseptic meningitis.
The key strengths of this study are the ability to identify population-based information in a nationwide cohort and link this across several data sources using the Danish CPR number. Using these tools, we were able to obtain a high retrieval rate of the relevant medical files and construct a suitable reference standard. Furthermore, we retrieved information during an 11-year period. The nationwide cohort with equal access to hospital services reduces referral bias. Our definition of encephalitis included confirmation of microbiological findings of HSV in the CSF and/or the acute involvement of brain parenchyma either by clinical symptoms or by brain imaging with findings in frontal or temporal regions characteristic of HSE.4 This is a highly reproducible definition, which ensures a strict confirmation of this disease. However, this definition might be too narrow and thereby contribute to an underestimation of the PPV. The sensitivity and specificity of PCR as a diagnostic technique for HSE have been estimated to be 98% and 94%, respectively.23 Thereby, we found it reasonable to rely solely on verification by microbiological data (if HSV-1 positive) without reviewing the medical records in these cases.
The low PPVs of the hospital diagnoses of HSE pose some potential problems in some analytic and incidence studies, perhaps requiring an alternative and/or additional validation algorithm. In this context, MiBa serves as a valuable resource of microbiological data from 2010 and onward. The authors recommend the use of MiBa in future validation studies also using microbiological data as part of the reference standard. However, the level of data quality needed for registry-based studies depends on the research question and study design.24 If data are used to compare incidence of HSE over time, the PPV should be stable over time.25 We found no significant changes in HSE coding validity during the 11-year study period but a tendency of an improved coding validity during the last 2 years of the study period. This may indicate improvement in the use of diagnostic tools with routine use of PCR instead of serological analyses. The gold standard for detecting HSV in CSF has been PCR throughout the study period. This was reflected by the fact that the annual number of incidents was stable and no decrease in the number of probable cases was observed. This is in contrast to a study by Kadambari et al reporting a significant increase in the laboratory-confirmed diagnoses of viral meningoencephalitis from 2004 to 2013.26
We found that the miscoding and thereby misclassification of HSE ICD-10 codes was substantial both with respect to etiology of encephalitis and the distinction between encephalitis and meningitis. More than half of the nonconfirmed cases had positive VZV in the CSF. Encephalitis due to VZV is more correctly contained by the ICD-10 code B02.0, and therefore, this is an example of miscoding by mixing of ICD-10 diagnosis codes. Moreover, we did not expect secondary diagnoses to have as high PPVs as primary diagnosis, since primary diagnosis should reflect the main reason for hospitalization. To improve the diagnosis coding of this disease, we recommend clinical guidelines for the distinction between encephalitis and meningitis emphasizing a combination of several diagnostic criteria as described by Granerod et al, which we also used in this study.14 Furthermore, more awareness and education regarding the diagnosis codes available as well as enhanced focus on the importance of precise diagnosis coding is recommended for both physicians and medical secretaries.
Conclusion
We found that the PPVs of the ICD-10 diagnosis coding for adult HSE in the DNPR were generally low. Therefore, ICD-10 diagnoses of HSE should be used with caution in epidemiological research and consolidated by microbiological data or information from medical records when possible.
Acknowledgments
The authors would like to thank doctors and secretaries at medical departments all over Denmark for their assistance with the retrieval of medical records. Furthermore, the authors would like to thank the Danish Microbiology Database (MiBa) Board of Representatives and departments of clinical microbiology at Aalborg University Hospital and Odense University Hospital for providing microbiological data. This study was funded by the Novo Nordisk Foundation, Denmark (application no 12083, grant area 460).
Disclosure
THM was supported by a grant from the Medical Research Council (DFF-4004-00047). The authors have no other conflicts of interest to declare in this work.
References
Granerod J, Ambrose HE, Davies NW, et al. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10(12):835–844. | ||
Whitley RJ. Herpes simplex virus infections of the central nervous system. Continuum (Minneap Minn). 2015;21(6):1704–1713. | ||
Whitley RJ, Soong SJ, Dolin R, Galasso GJ, Ch’ien LT, Alford CA. Adenine arabinoside therapy of biopsy-proved herpes simplex encephalitis. National Institute of Allergy and Infectious Diseases collaborative antiviral study. N Engl J Med. 1977;297(6):289–294. | ||
Whitley RJ. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 2006;71(2–3):141–148. | ||
Riancho J, Delgado-Alvarado M, Sedano MJ, Polo JM, Berciano J. Herpes simplex encephalitis: clinical presentation, neurological sequelae and new prognostic factors. Ten years of experience. Neurol Sci. 2013;34(10):1879–1881. | ||
Hjalmarsson A, Blomqvist P, Sköldenberg B. Herpes simplex encephalitis in sweden, 1990-2001: incidence, morbidity, and mortality. Clin Infect Dis. 2007;45(7):875–880. | ||
Aurelius E, Johansson B, Sköldenberg B, Forsgren M. Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or 2 as determined by type-specific polymerase chain reaction and antibody assays of cerebrospinal fluid. J Med Virol. 1993;39(3):179–186. | ||
Sørensen HT, Lash TL, Rothman KJ. Beyond randomized controlled trials: a critical comparison of trials with nonrandomized studies. Hepatology. 2006;44(5):1075–1082. | ||
Pedersen CB, Gotzsche H, Møller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53(4):441–449. | ||
Frank L. Epidemiology. When an entire country is a cohort. Science. 2000;287(5462):2398–2399. | ||
Schmidt M, Schmidt SA, Sandegaard JL, Ehrenstein V, Pedersen L, Sørensen HT. The Danish National Patient Registry: a review of content, data quality, and research potential. Clin Epidemiol. 2015;7:449–490. | ||
Voldstedlund M, Haarh M, Mølbak K, MiBa Board of Representatives. The Danish Microbiology Database (MiBa) 2010 to 2013. Euro Surveill. 2014;19(1):20667. | ||
Sundhedsdatastyrelsen. Koder der lukkes og erstattes af anden kode. [Codes that are closed and replaced by other code]. Available from: http://sundhedsdatastyrelsen.dk/da/rammer-og-retningslinjer/om-klassifikationer/sks-klassifikationer/klassifikation-sygdomme/kodeaendringer. Accessed May 10, 2016. Danish. | ||
Granerod J, Cunningham R, Zuckerman M, et al. Causality in acute encephalitis: defining aetiologies. Epidemiol Infect. 2010;138(6):783–800. | ||
Persson A, Bergström T, Lindh M, Namvar L, Studahl M. Varicella-zoster virus CNS disease--viral load, clinical manifestations and sequels. J Clin Virol. 2009;46(3):249–253. | ||
Fagan T. Exact 95% confidence intervals for differences in binomial proportions. Comput Biol Med. 1999;29(1):83–87. | ||
Blichert-Hansen L, Nielsson MS, Nielsen RB, Christiansen CF, Nørgaard M. Validity of the coding for intensive care admission, mechanical ventilation, and acute dialysis in the danish national patient registry: a short report. Clin Epidemiol. 2013;5:9–12. | ||
Heden KE, Jensen AO, Farkas DK, Norgaard M. Validity of a procedure to identify patients with chronic idiopathic thrombocytopenic purpura in the danish national registry of patients. Clin Epidemiol. 2009;1:7–10. | ||
Holland-Bill L, Xu H, Sorensen HT, et al. Positive predictive value of primary inpatient discharge diagnoses of infection among cancer patients in the Danish National Registry of Patients. Ann Epidemiol. 2014;24(8):593–597, 597.e1–597.e18. | ||
Jespersen CG, Borre M, Norgaard M. Validity of the recorded codes of gonadotropin-releasing hormone agonist treatment and orchiectomies in the Danish National Patient Registry. Clin Epidemiol. 2012;4:145–149. | ||
Brenner H, Savitz DA. The effects of sensitivity and specificity of case selection on validity, sample size, precision, and power in hospital-based case-control studies. Am J Epidemiol. 1990;132(1):181–192. | ||
Heininger U, Seward JF. Varicella. Lancet. 2006;368(9544):1365–1376. | ||
Lakeman FD, Whitley RJ. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group. J Infect Dis. 1995;171(4):857–863. | ||
Sorensen HT, Sabroe S, Olsen J. A framework for evaluation of secondary data sources for epidemiological research. Int J Epidemiol. 1996;25(2):435–442. | ||
Nørgaard M, Skriver MV, Gregersen H, Pedersen G, Schonheyder HC, Sorensen HT. The data quality of haematological malignancy ICD-10 diagnoses in a population-based hospital discharge registry. Eur J Cancer Prev. 2005;14(3):201–206. | ||
Kadambari S, Okike I, Ribeiro S, et al. Seven-fold increase in viral meningo-encephalitis reports in England and Wales during 2004-2013. J Infect. 2014;69(4):326–332. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.