Back to Journals » Cancer Management and Research » Volume 11
Upregulation Of circMMP9 Promotes Osteosarcoma Progression Via Targeting miR-1265/CHI3L1 Axis
Authors Pan G, Hu T, Chen X, Zhang C
Received 6 August 2019
Accepted for publication 5 October 2019
Published 29 October 2019 Volume 2019:11 Pages 9225—9231
DOI https://doi.org/10.2147/CMAR.S226264
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Guangjie Pan,1 Ting Hu,1 Xuewu Chen,1 Chao Zhang2
1Department of Orthopedics, Wenzhou Central Hospital, Wenzhou 325000, People’s Republic of China; 2Department of Orthopedics, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Correspondence: Chao Zhang
Department of Orthopedics, The First Affiliated Hospital of Wenzhou Medical University, Shangcai Village, Ouhai District, Wenzhou 325000, People’s Republic of China
Tel +86 15958710270
Email [email protected]
Background: Osteosarcoma (OS) is a very aggressive cancer. Nevertheless, how circular RNA (circRNA) contributes to OS progression remains unclear. Here, we aimed to research the functions of circMMP9 in OS progression.
Methods: Gene expression was determined via qRT-PCR. siRNA was used to knock down circMMP9. Proliferation was analyzed using CCK8 and colony formation assays. Migration and invasion were measured using Transwell assay.
Results: circMMP9 was overexpressed in cancer tissues. Overexpressed circMMP9 was correlated with advanced tumor stage and predicted poor prognosis. circMMP9 knockdown exhibited a tumor-suppressive phenotype via suppressing proliferation, migration and invasion. Besides, decreased circMMP9 level promoted OS cellular apoptosis. Mechanistically, circMMP9 was shown to be located in the cytoplasm and sponge miR-1265. Furthermore, miR-1265 directly targeted CHI3L1. CHI3L1 levels were modulated by circMMP9/miR-1265 axis. Rescue experiments indicated that circMMP9 contributes to OS development through the miR-1265/CHI3L1 pathway.
Conclusion: Our findings provide a novel insight about how circRNA regulates OS progression.
Keywords: circMMP9, miR-1265, CHI3L1, osteosarcoma, progression
Introduction
(OS) is one of the most aggressive bone tumors among young people.1 OS causes the third largest number of deaths worldwide among children.2 Although therapeutic strategies against OS have been developed in recent decades, patients with metastasis still display poor outcomes.3 Thus, it is important to explore the molecular mechanism of OS development and progression and screen out novel therapeutic targets for OS treatment.
Circular RNA (circRNA) is a type of noncoding RNAs characterized by the existence of a close loop. In the past years, circRNAs have been reported to exert critical functions in the initiation and progression of various cancers.4 Many evidences indicate that a lot of circRNAs are aberrantly expressed in tumor tissues and participate in the regulation of proliferation, migration and invasion.5,6 For instance, circSLC8A1 is downregulated in bladder cancer tissues and suppresses cancer development via upregulating PTEN and restraining miR-130b/miR-494 activity.7 circPTPRA level is decreased in non-small cell lung cancer (NSCLC) and it inhibits tumor metastasis through interacting miR-96-5p.8 Circ_0020123 is overexpressed in NSCLC tissues and contributes to cancer development via sponging miR-488.9 Additionally, several evidences also show many circRNAs are dysregulated in OS tissues.10,11 circTADA2A was identified to promote OS cell migration and invasion while hsa_circ_0004674 increases the chemo-resistance of OS cells.10,11 Thus, circRNA may also have essential roles in OS and more investigations are required.
CircMMP9 has been shown to promote glioblastoma development.12 CircMMP9 is highly expressed in GBM cells and accelerates proliferation, migration and invasion.12 We also observed circMMP9 upregulation in OS tissues. However, its role in OS remains unknown. In this study, we showed circMMP9 was overexpressed in OS tissues and predicted poor prognosis. CircMMP9 knockdown suppressed proliferation, migration and invasion. Mechanistically, circMMP9 was found to sponge miR-1265 and upregulate CHI3L1. In conclusion, our results reveal that circMMP9/miR-1265/CHI3L1 axis modulates OS progression.
Materials And Methods
Patient Samples
Fifty-one pairs of OS tissues and adjacent normal controls were obtained from Wenzhou Central Hospital. Prior to surgery, all patients received no chemotherapy or radiotherapy. Tissues were stored in liquid nitrogen. This study was approved by the Ethics Committee of Wenzhou Central Hospital. Experiments involving human tissues were conducted in accordance with the Declaration of Helsinki. Written informed consents were provided by the participants.
Cell Culture
Human OS cell lines (U2OS, HOS, Saos2 and MG63 cells) and osteoblast cell line hFOB 1.19 were from Shanghai Academy of Sciences. Cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100 μg/mL) and maintained at 37°C with 5% CO2.
RNA Transfection
siRNAs against circMMP9 (5ʹ-GUGGAGGCGCAGAUGGUGGAU-3ʹ) or CHI3L1 (5ʹ-AUGUAAGACUCGGGAUUAGUA-3ʹ) and negative controls were purchased from GenePharma (Shanghai, China). siRNAs (50 nM), miRNA mimics (50 nM) and miRNA inhibitors (50 nM) were transfected into cells using Lipofectamine™ RNAiMAX (Invitrogen) based on the manufacturer’s protocols. After transfection for 48 h, transfection efficiency was validated by qRT-PCR as described below.
RNA Extraction And Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were isolated using TRIzol reagent (Invitrogen) and the RNA quality was monitored using NanoDrop 2000. Then, total RNA was reversely transcribed into cDNA using a reverse transcription kit (Takara, Tokyo, Japan). Then, qPCR was completed using SYBR-Green PCR kit (QIAGEN, Hilden, Germany) on an ABI 7300 Fast Real-Time PCR system. Relative expression was normalized to GAPDH and calculated based on the 2−ΔΔCt method. Primer sequences were as follows: circMMP9 (Forward, 5ʹ-CCAGTTACAAGT TTAGGGCTGT-3ʹ and reverse, 5ʹ-TGTCTCCATTTGCTTCTTCTTCA-3ʹ), CHI3L1 (Forward, 5ʹ-CCCAACCTGAAGACTCTCTTG-3ʹ and reverse, 5ʹ-CCAAGATAGCCTCCAACACC-3ʹ) and GAPDH (Forward, 5ʹ-GCCATCACAGCAACACAGAA-3ʹ and reverse, 5ʹ-GCCATACCAGTAAGCTTG CC-3ʹ).
CCK8 Assay
Transfected OS cells were seeded into the 96-well plates and cultured for indicated days. Then, 20 μl CCK8 solution was added and incubated for 4 hrs, followed by detection of absorbance at 450 nm using a microplate reader at 490 nm (Thermo Fisher, Massachusetts, USA).
Transwell Assay
The migration and invasion were determined by using Transwell plates (8 μm pore membrane, Costar, Cambridge, MA, USA). The detailed protocols were described previously.13 In brief, 2×104 cells per well were seeded into the serum-free medium of the upper chamber. The lower chamber was filled with complete medium. After cultured for 48 hrs, the migrated or invaded cells were fixed, stained and counted.
Colony Formation Assay
Transfected OS cells were seeded into the 6-well plates and maintained for 14 days. Then, the colonies were fixed and stained with 0.5% crystal violet. The numbers of colonies were finally counted using the ImageJ software.
Dual-Luciferase Reporter Assays
The sequences of circMMP9 or CHI3L1 were inserted into the pGL3 luciferase vector (Promega, USA). For luciferase reporter assay, the luciferase reporter vector and miR-1265 mimics or controls were co-transfected into U2OS cells. Forty-eight hours later, the luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instruction.
Cytoplasmic And Nuclear RNA Isolation
The cytoplasmic and nuclear fractions were isolated using PARIS Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturers’ instructions, followed by qRT-PCR analysis.
Caspase Activity Assay
The activities of caspase-3/7 were measured using caspase-3/7 activity kit following the manufacturers’ instructions.
Statistical Analysis
Statistical analysis was completed using Graphpad Prism 6 software and results were expressed as mean ± standard deviation. Comparisons between experimental group and control group were performed using Student’s t-test or one-way ANOVA. qRT-PCR was used to determine circMMP9 low or high expression in tissues. Clinical data were analyzed using Fisher’s exact test, Kaplan–Meier curves with log-rank test. A P-value (<0.05) was considered as statistically significant.
Results
circMMP9 Was Overexpressed In OS
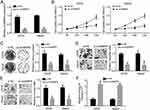
To investigate the correlation between circMMP9 expression and OS development, its expression in OS tissues was analyzed. circMMP9 level was higher in OS tissues than that in adjacent normal ones (P<0.05; Figure 1A). Besides, circMMP9 expression was increased in OS tissues with larger tumor size and advanced stages (P<0.05; Figure 1B and C). Moreover, we found that circMMP9 expression was positively correlated with metastasis, but not gender or age (Table 1). Then, circMMP9 expression in OS cell lines was further researched. As shown, increased circMMP9 expression was observed in OS cell lines compared to hFOB1.19 cells (Figure 1D). OS samples were divided into two subgroups based on circMMP9 level. Kaplan–Meier curve indicated that circMMP9 overexpression predicted a low survival rate (Figure 1E). Finally, we analyzed the localization of circMMP9 in OS cells and found that circMMP9 was expressed in cytoplasm of U2OS cells (Figure 1F).
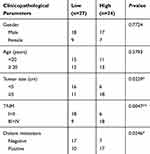 |
Table 1 Association Between circMMP9 Expression And Clinicopathological Parameters In OS |
Knockdown Of circMMP9 Showed An Anti-Tumor Phenotype
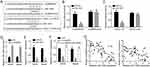
To investigate the potential roles of circMMP9, we knocked down it in U2OS and Saos2 cells (Figure 2A). CCK8 and colony formation assay obviously indicated that circMMP9 knockdown inhibited the cellular viability and colony numbers (Figure 2B and C), suggesting circMMP9 depletion suppressed proliferation. Transwell assay showed that the cell numbers of migration and invasion were significantly declined after circMMP9 silencing (Figure 2D and E). Moreover, we found that circMMP9 knockdown increased the activity of Caspase-3/7 (Figure 2F), indicating circMMP9 knockdown promoted apoptosis.
circMMP9 Modulated The miR-1265/CHI3L1 Axis
Then, we analyzed the potential mechanism using bioinformatics methods. Through CircInteractome and TargetScan tools, we found that circMMP9 may be a sponge for miR-1265 and miR-1265 may directly target CHI3L1. We identified their potential interactive sites and mutated the sites (*P<0.05; Figure 3A). Luciferase reporter assay showed that miR-1265 mimic suppressed the activity of circMMP9-wt or CHI3L1-wt reporter (*P<0.05; Figure 3B and C), demonstrating their interactions. Additionally, circMMP9 knockdown led to increased expression of miR-1265 (*P<0.05; Figure 3D). And miR-1265 knockdown inhibited the mRNA level of CHI3L1 (*P<0.05; Figure 3E). Moreover, CHI3L1 expression was regulated by circMMP9 mediated inhibition of miR-1265 (*P<0.05; Figure 3F). Of note, we observed that miR-1265 expression was negatively correlated with circMMP9 and CHI3L1 in OS tissues (Figure 3G).
circMMP9/miR-1265/CHI3L1 Axis Contributed To OS Progression
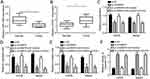
Through qRT-PCR, we found that miR-1265 was downregulated in OS tissues while CHI3L1 was upregulated (Figure 4A and B), suggesting their potential roles in OS progression. To determine the roles of circMMP9/miR-1265/CHI3L1 axis, we performed rescue assays. We found that miR-1265 inhibitors rescued the proliferation, migration, invasion and apoptosis in U2OS and Saos2 cells with circMMP9 knockdown (Figure 4C–F). Moreover, knockdown of CHI3L1 further suppressed the proliferation, migration and invasion while promoting apoptosis in U2OS and Saos2 cells transfected with si-circMMP9 and miR-1265 inhibitors (Figure 4C–F). Taken together, circMMP9/miR-1265/CHI3L1 axis plays a key role in regulating OS progression.
Discussion
Until now, the pathological mechanism in OS is still unclear. Several evidences indicate that the dysregulation of oncogenes and anti-tumor genes is correlated with OS development and progression.2 In this research, we explored how circMMP9 regulates OS progression. We showed circMMP9 expression was upregulated in OS tissues and correlated with advanced stage and metastasis. CircMMP9 upregulation predicted poor prognosis. Knockdown of circMMP9 inhibited the malignant behaviors of OS cells. And circMMP9 was identified to sponge miR-1265, causing upregulation of CHI3L1.
circRNAs are a type of novel noncoding RNAs. The study about their function is emerging in recent years. Although researches about circRNAs are rather limited, their important functions have been widely demonstrated, especially in tumor biology.14 In OS, several important circRNAs have been identified. For example, circ-0001785 promotes OS growth and metastasis via inhibiting miR-1200 to increase HOXB2 level.15 CircTADA2A enhances OS cell proliferation and invasion through restricting miR-203a and upregulating CREB3.10 Additionally, circRNA CDR1as has been identified to increase OS growth and serve as a diagnostic marker.16 In this study, we identified circMMP9 as a tumor driver in OS. We showed that circMMP9 was overexpressed in OS tissues and predicted poor prognosis. CircMMP9 knockdown suppressed the proliferation, migration and invasion in vitro. Our findings suggest circMMP9 might be a novel therapeutic target for OS intervention.
The circRNA-miRNA-mRNA network has been indicated to participate in the regulation of tumorigenesis.17,18 circRNA could interact with the response element of miRNA and inhibit the activity of miRNA.19 For instance, circRNA cTFRC is the sponge for miR-107 to participate in bladder cancer development.20 CircPSMC3 interacts with miR-296-5p to repress gastric cancer development.21 Hsa-circ-0014359 sponges miR-153 to increase glioma tumorigenesis.22 In our study, we identified circMMP9 as the sponge for miR-1265. We then demonstrated their interaction via luciferase reporter assay and showed that miR-1265 level was inhibited by circMMP9. MiR-1265 is a poorly explored miRNA. A research shows that miR-1265 inhibits gastric cancer cell growth and promotes apoptosis.23 Whether miR-1265 regulates OS progression remains unclear. Our data showed that miR-1265 expression was downregulated in OS tissues and suppressed the proliferation, migration and invasion of OS cells, suggesting miR-1265 as a novel tumor suppressor in OS. Afterwards, we further identified the target of miR-1265. We showed that miR-1265 directly bound to the 3ʹ UTR of CHI3L1 mRNA and inhibited its expression. The oncogenic roles of CHI3L1 have been well studied in several tumors, such as hepatocellular carcinoma, ovarian cancer and cervical cancer.24–26 However, the function of CHI3L1 in OS is not clear. We also indicated that CHI3L1 expression was upregulated in OS tissues. And CHI3L1 expression was modulated by circMMP9/miR-1265 axis. Finally, we demonstrated that CHI3L1 knockdown inhibited the proliferation, migration and invasion while enhancing apoptosis in OS cells. Our results identified CHI3L1 as a new oncogene in OS.
In conclusion, our research identified a novel mechanism that circMMP9/miR-1265/CHI3L1 axis contributes to OS cell proliferation, migration and invasion. However, there are some limitations in our work. For example, it would be best to run a Cox-proportionate Hazard Model analysis to see if there would be any difference and how the expression can affect survival when the TNM staging is taken into consideration as a potential confounding factor.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi:10.1016/j.ocl.2015.08.022
2. Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–19. doi:10.1097/01.JAA.0000541477.24116.8d
3. He JP, Hao Y, Wang XL, et al. Review of the molecular pathogenesis of osteosarcoma. Asian Pac J Cancer Prev. 2014;15(15):5967–5976. doi:10.7314/apjcp.2014.15.15.5967
4. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
5. Liu W, Zhang J, Zou C, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43(3):969–985. doi:10.1159/000481650
6. Wang C, Ren M, Zhao X, Wang A, Wang J. Emerging roles of circular RNAs in osteosarcoma. Med Sci Monit. 2018;24:7043–7050. doi:10.12659/MSM.912092
7. Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111. doi:10.1186/s12943-019-1010-6
8. Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi:10.1016/j.ebiom.2019.05.032
9. Wan J, Hao L, Zheng X, Li Z. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515(2):303–309. doi:10.1016/j.bbrc.2019.05.158
10. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1010-6
11. Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin Z, Jian-Ping H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327–1346. doi:10.2217/epi-2018-0023
12. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. doi:10.1186/s12943-018-0911-0
13. Yanbin Z, Jing Z. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Commun. 2019. doi:10.1016/j.bbrc.2019.05.182
14. Qiu M, Xia W, Chen R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi:10.1158/0008-5472.CAN-17-2808
15. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. 2019;18(11):1281–1291. doi:10.1080/15384101.2019.1618127
16. Xu B, Yang T, Wang Z, Zhang Y, Liu S, Shen M. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880. doi:10.2147/CMAR.S178213
17. Liu H, Liu Y, Bian Z, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 2018;17(1):151. doi:10.1186/s12943-018-0902-1
18. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi:10.1186/s12943-017-0719-3
19. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
20. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27. doi:10.1186/s12943-019-1010-6
21. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18(1):25. doi:10.1186/s12943-019-1010-6
22. Shi F, Shi Z, Zhao Y, Tian J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510(4):614–620. doi:10.1016/j.bbrc.2019.02.019
23. Xu Z, Li Z, Wang W, et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett. 2019;449:226–236. doi:10.1016/j.canlet.2019.02.026
24. Qiu QC, Wang L, Jin SS, et al. CHI3L1 promotes tumor progression by activating TGF-beta signaling pathway in hepatocellular carcinoma. Sci Rep. 2018;8(1):15029. doi:10.1038/s41598-018-33239-8
25. Lin HW, Chiang YC, Sun NY, et al. CHI3L1 results in poor outcome of ovarian cancer by promoting properties of stem-like cells. Endocr Relat Cancer. 2019;26(1):73–88. doi:10.1530/ERC-18-0300
26. Ngernyuang N, Shao R, Suwannarurk K, Limpaiboon T. Chitinase 3 like 1 (CHI3L1) promotes vasculogenic mimicry formation in cervical cancer. Pathology. 2018;50(3):293–297. doi:10.1016/j.pathol.2017.09.015
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.